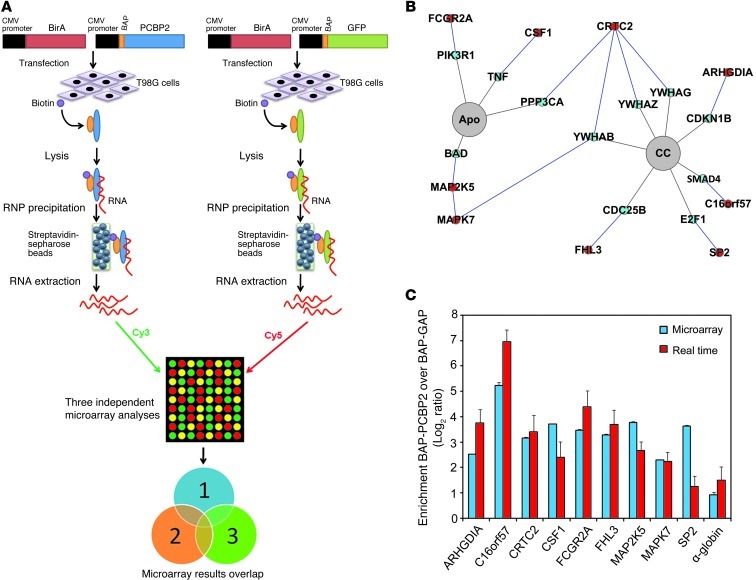
Figure 4. RIP-ChIP analysis of PCBP2 and identification of preferentially associated mRNAs.
(A) Schematic outline of the RIP-ChIP analysis to identify PCBP2-associated mRNAs. T98G cells were transfected transiently with BAP-tagged constructs. BAP-tagged proteins were biotinylated in vivo by the cotransfected hBirA enzyme. RNPs were recovered via precipitation with streptavidin-sepharose beads. Finally, RNAs were purified and analyzed on microarrays. The BAP-GFP control was used in 3 independent sets of experiments. (B) Detection of the “protein-protein interaction” and “pathway” relationships between the proteins encoded by the PCBP2-associated mRNAs via the CSPN. In our pathways of interest (highlighted in gray), we found 9 proteins encoded by the PCBP2-associated mRNAs (highlighted in red) and their interacting protein partners (highlighted in blue). (C) Real-time PCR validation of PCBP2-associated mRNAs identified by RIP-ChIP. We selected 9 mRNAs to be validated by real-time PCR. We also verified the enrichment of α-globin, a previously identified PCBP2 target, in the BAP-PCBP2–associated mRNA population. Blue and red bars represent the enrichment observed with the microarrays and real-time PCR, respectively. Apo, apoptosis; CC, cell cycle.