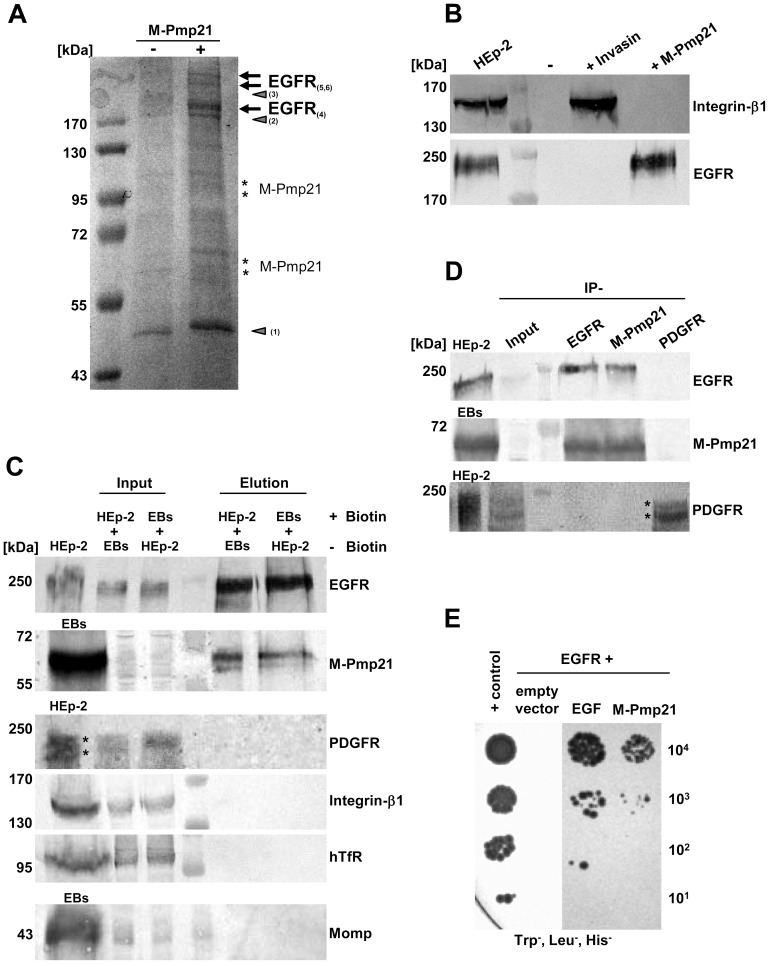
Figure 2. Identification of the EGF receptor as an interaction partner for the C. pneumoniae adhesin Pmp21.
(A) Electrophoretic analysis of fractions eluted from a NeutrAvidin column. HEp-2 cells were incubated with (+) or without (−) biotinylated M-Pmp21 or invasin (data for invasin not shown), and processed as described in Experimental Procedures. Arrows mark bands in which EGFR was identified by MALDI-MS (see Fig. S2B; bands 4–6). Other major bands (triangles) were identified as actin (1) and pleckstrin (2); no protein was detected in band 3. Bands marked with asterisks represent the recombinant M-Pmp21 as shown by immunoblotting (data not shown). (B) Fractions from (A) were probed with anti-integrin-β1 and anti-EGFR antibodies. (C) Affinity purification of the EGFR-Pmp21 complex from surface-biotinylated HEp-2 cells incubated with non-biotinylated C. pneumoniae EBs, or vice versa, for 60 min at 37°C. After crosslinking the biotinylated surface protein complexes were bound to a NeutrAvidin column. After crosslink removal the eluted proteins were identified by immunoblot analysis with specific antibodies against EGFR, M-Pmp21, PDGFR, integrin-β1, hTfR and Momp. Equal amounts of input and elution samples were loaded onto the SDS-PAGE. (D) Coimmunoprecipitation of the EGFR-Pmp21 complex from HEp-2 monolayers incubated with purified C. pneumoniae EBs (MOI 5) for 60 min. The Pmp21-EGFR complex was precipitated using EGFR-, M-Pmp21- and PDGFR-specific antibodies as indicated, and probed after SDS-PAGE and immunoblotting. Stars mark the 2 main PDGFR protein species recognized. (E) Interaction of EGFR and M-Pmp21 shown by Y2H analysis. Serial dilutions (101–104) of yeast cells expressing EGFR/EGF or EGFR/M-Pmp21 were patched on low-stringency selection medium (Leu−, Trp−, His−). + control: SV40 LTA/p53.