Figure 1. Retrohoming pathway of Ll.LtrB intron lariat RNA in bacteria.
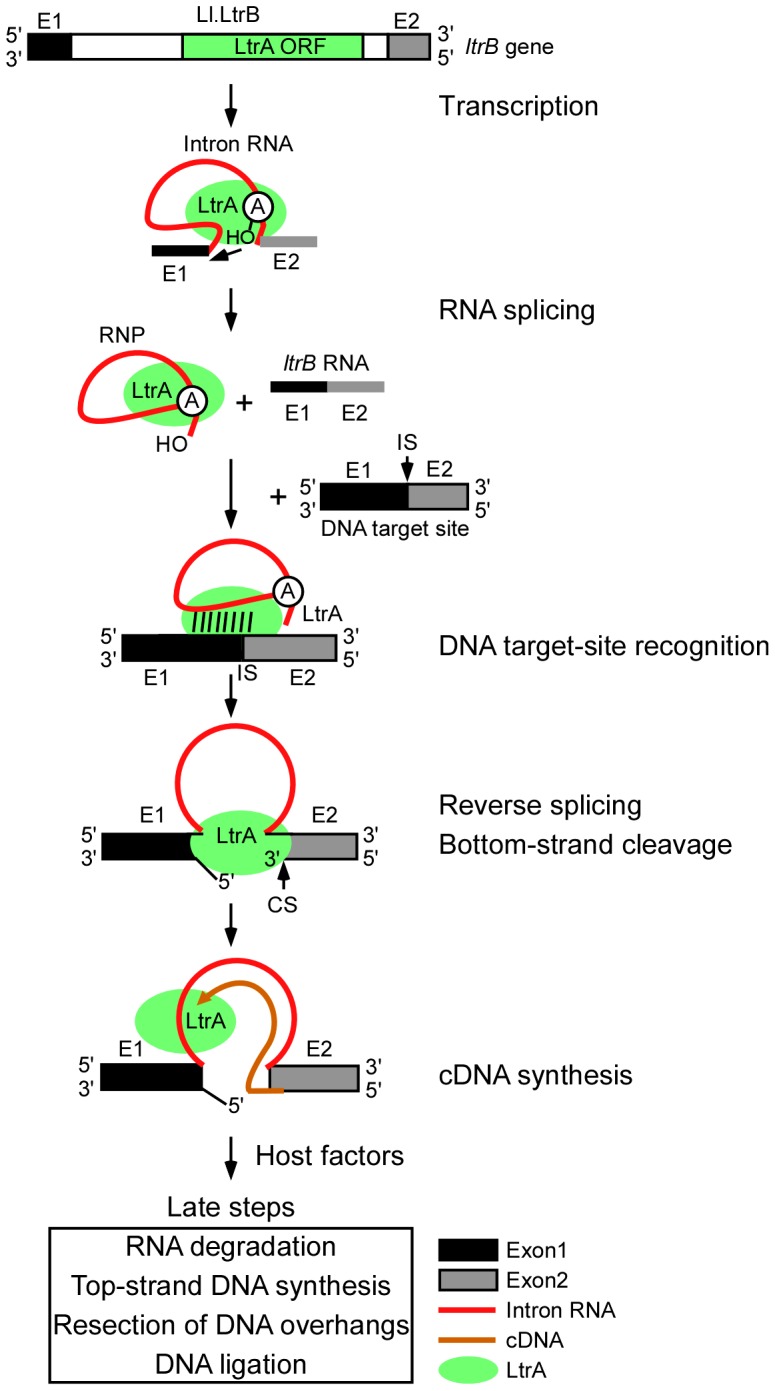
The Ll.LtrB intron, found in a relaxase gene (ltrB) in an L. lactis conjugative element, encodes a multi-functional RT (LtrA protein) with RT, RNA splicing, DNA-binding, and DNA endonuclease activities. Transcription of the ltrB gene yields a precursor RNA containing the intron flanked by 5′ and 3′ exons (E1 and E2, respectively). LtrA is translated from within the intron using its own Shine-Dalgarno sequence and then binds to the intron in the precursor RNA to promote formation of the catalytically active RNA structure for RNA splicing. RNA splicing occurs via two sequential RNA-catalyzed transesterification reactions that are initiated by nucleophilic attack of the 2′ OH of a branch point A-residue near the 3′ end of the intron at the 5′-splice site and results in ligated ltrB exons and an excised intron lariat RNA with a 2′-5′ phosphodiester linkage. After splicing, LtrA remains tightly bound to the excised intron lariat RNA in an RNP. RNPs initiate retrohoming by recognizing a DNA target site (the ligated ltrB E1–E2 sequence), using both the IEP and base pairing of the intron RNA. The intron RNA then inserts via reversal of the two transesterification reactions used for RNA splicing (referred to as “full reverse splicing”) into the intron-insertion site (IS) at the ligated-exon junction in the top strand of the DNA target site. LtrA uses its DNA endonuclease activity to cleave the bottom strand at a site (CS) between positions +9 and +10 of E2 and uses the 3′ DNA end at the cleavage site as a primer for reverse transcription of the inserted intron RNA. The resulting intron cDNA is then integrated into the genome by host enzymes in late steps that minimally include degradation of the intron RNA template strand, second (top)-strand DNA synthesis, resection of DNA overhangs, and sealing of DNA strand nicks [12].
