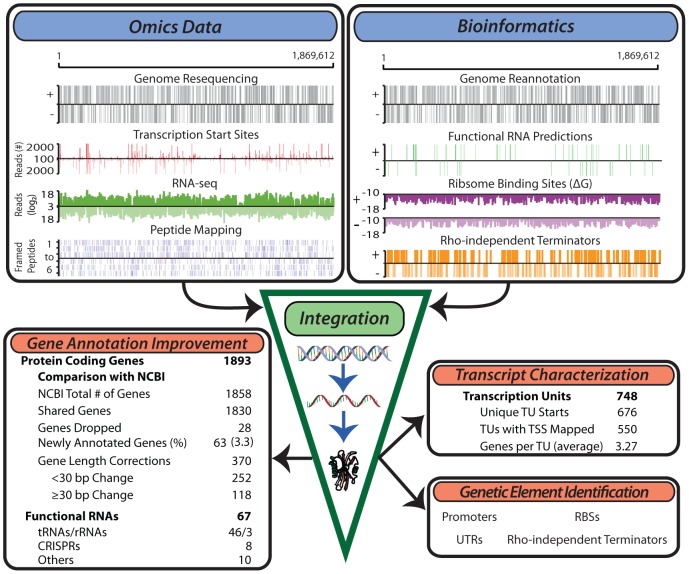
Figure 1. Generation of multiple genome-scale datasets integrated with bioinformatics predictions reveals the genome organization.
Experimental data generated for the study of the T. maritima genome include genome resequencing, TSS determination, RNA-seq, tiling arrays (not shown) and LC-MS/MS peptide mapping (top left). Bioinformatics approaches used include genome re-annotation, functional RNA prediction, ribosome binding site energy calculations, and determination of intrinsic terminators (top right). Integration of these distinct datasets involves normalization and quantification to genomic coordinates. This experimentally anchors gene annotation improvements, defines the TU architecture, identifies non-coding RNAs and serves as a basis for the identification of additional genetic elements such as promoters and ribosome binding sites.