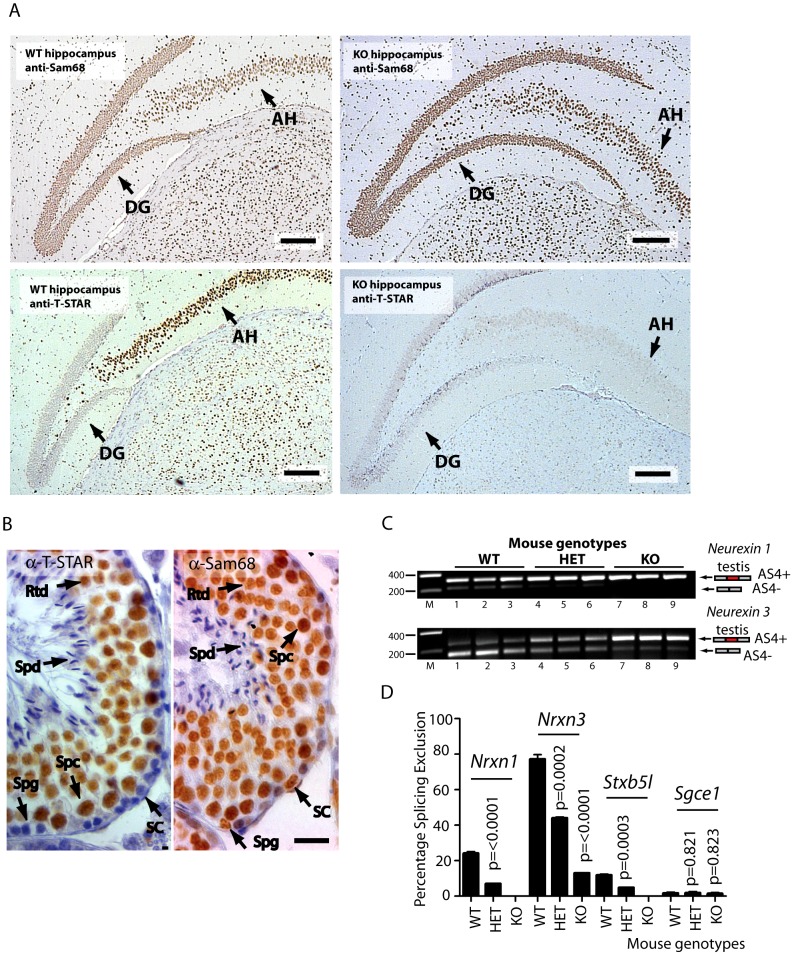
Figure 7. Nrxn exon AS4 alternative splicing control is dependent on the physiological expression of T-STAR protein even though Sam68 is co-expressed.
(A) Immunolocalisation of T-STAR and Sam68 proteins in the mouse hippocampus from wild type or knockout mouse brains (Abbreviations: DG - Dentate Gyrus; and AH -Ammon's Horn). The scale bar is equivalent to 20 µm). (B) Immunolocalisation in the mouse testis. Paraffin embedded adult mouse testis sections were stained with affinity purified antibodies raised against T-STAR or Sam68 (brown staining), and counterstained with haematoxylin (blue). Abbreviations: Spg –spermatogonia; Spc –spermatocyte; Rtd –round spermatid; Spd –elongating spermatid; SC –Sertoli cell. The size bar corresponds to 20 µM. (C) Levels of Nrxn1 and Nrxn3 AS4 alternative splice isoforms in the testes of different mouse genotypes (n = 3 mice of each genotype) measured by RT-PCR and agarose gel electrophoresis. (D) Quantification of Percentage Splicing Exclusion in the testes of different mouse genotypes using capillary gel electrophoresis (n = 3 mice of each genotype: wild type mice Khdrbs3+/+ (abbreviated WT) Khdrbs3+/− mice (abbreviated HET) and Khdrbs3−/− mice (abbreviated KO). The p values were calculated using unpaired t tests, to determine the significance of the difference between percentage splicing exclusion levels in the wild type versus either the heterozygous Khdrbs+/− mice (HET); or wild type versus the homozygous Khdrbs3−/− (KO) mice. The standard error of the mean is shown as an error bar.