Abstract
We demonstrate that alignment of a structured peptide or small protein solubilized in mixed phospholipid:detergent micelles or bicelles, when embedded in a compressed gel or liquid crystalline medium, can be altered by either changing the phospholipid aggregate shape, charge, or both together. For the hemagglutinin fusion peptide solubilized in bicelles, we show that bicelle shape and charge do not change its helical hairpin structure but impact its alignment relative to the alignment medium, both in charged compressed acrylamide gel and in liquid crystalline d (GpG). The method can be used to generate sets of residual dipolar couplings (RDCs) that correspond to orthogonal alignment tensors, and holds promise for high-resolution structural refinement and dynamic mapping of membrane proteins.
Keywords: bicelle, dipolar coupling, fusion peptide, NMR, orthogonal alignment, Saupe matrix
Introduction
Residual dipolar couplings (RDCs) in biomolecules are particularly useful NMR parameters for the study of molecular structure and dynamics (Prestegard et al. 2000; Bax 2003). They can be observed by solution NMR when the molecules are aligned very weakly relative to the magnetic field. Such alignment can be induced by a variety of dilute lyotropic liquid crystalline suspensions, including bicelles (Tjandra and Bax 1997) and polyethylene glycol based bilayers (Ruckert and Otting 2000), or by dissolving the protein in a liquid crystalline suspension of filamentous phages (Clore et al. 1998; Hansen et al. 1998). Alternatively, studying the protein in compressed hydrogels (Sass et al. 2000; Tycko et al. 2000) has proven to be a popular and robust method, also applicable to detergent-solubilized proteins. Alignment can also be obtained through paramagnetism of a protein, either by using a natural metal binding site (Tolman et al. 1995; Bertini et al. 2004) or by linking the protein to a metal-chelating tag (Wohnert et al. 2003; Su and Otting 2010).
RDCs report on the time-averaged magnetic dipole-dipole interactions, which depend on the averaged internuclear vector orientation in a laboratory frame. They are commonly used for refining biomolecular structures at improved resolution (Cornilescu et al. 1998; Clore et al. 1999; Ulmer et al. 2003; Bertini et al. 2009), for (cross-)validating the accuracy of a structure (Cornilescu et al. 1998; Clore and Garrett 1999), and for mapping the amplitude and directions of motions that reorient bond vectors (Meiler et al. 2001; Bertini et al. 2004; Blackledge 2005; Tolman and Ruan 2006).
In the case of a static protein model, a single RDC confines the orientation of its corresponding internuclear vector to the surfaces of two opposing cones. This orientational degeneracy can be partially lifted by measuring the RDC of the same internuclear vector under a condition where the protein is aligned in a different orientation with respect to the alignment medium (Ramirez and Bax 1998; Al-Hashimi et al. 2000), or by measuring multiple RDCs within a fragment of known, fixed geometry, such as the peptide bond (Mueller et al. 2000; Zidek et al. 2003; Hus et al. 2008).
Residual molecular alignment of a rigid molecule is described by a symmetric and traceless Saupe matrix (Saupe and Englert 1963; Losonczi et al. 1999; Bax et al. 2001) containing five independent elements, and up to five "orthogonal" alignment orientations potentially can be generated. With experimental RDCs for all five of these orthogonal alignments available, model-free information on the amplitude and direction of internal dynamics of the internuclear vector orientations can be obtained, provided the structure and dynamics are not impacted by the alignment process (Meiler et al. 2001; Briggman and Tolman 2003; Hus et al. 2003; Yao et al. 2008). Measurements under as few as three orthogonal alignments suffice to resolve the orientational degeneracy for the static case (Ruan et al. 2008; Yao et al. 2008). In practice, orthogonal alignments are rarely achieved, but any pair of linearly independent RDC measurements can be decomposed into two orthogonal sets, for which the normalized scalar product of the corresponding alignment tensors equals zero (Sass et al. 1999). Linearly independent RDC datasets can be generated by changing the nature of the interaction between the biomolecule of interest and the alignment medium. This can be achieved by changing the chemical nature of the liquid crystal or hydrogel, either by "electrostatic doping" (Ramirez and Bax 1998; Shortle and Ackerman 2001; Meier et al. 2002), or by combining data recorded in different liquid crystalline or otherwise ordered media (Sass et al. 1999; Peti et al. 2002). Alternatively, the alignment orientation can be changed by modifying the size (Zhang et al. 2006) or surface charge distribution (Yao and Bax 2007) of the biomolecule of interest, by altering the site where a paramagnetic tag is attached to the protein (Rodriguez-Castaneda et al. 2006), or by using different lanthanide ions chelated to the tag (Kamen et al. 2007).
In this study, we demonstrate a simple method for changing the orientation of the small, highly lipophilic protein domain composed of the first 23 residues of the second subunit of influenza hemagglutinin, HAfp23 (Lorieau et al. 2010), solubilized in small bicelles. Obtaining unique RDC datasets for membrane proteins is particularly challenging due to the paucity of liquid crystalline alignment media that are compatible with detergents and phospholipids. Detergents destructively interfere with most protein-compatible liquid crystalline suspensions, although two DNA-based media are less sensitive to this limitation. These include engineered DNA nanotubes (Douglas et al. 2007), as well as suspensions of the dinucleotide d (GpG), which self-assembles to form long quadruplex columns (Lorieau et al. 2008).
We demonstrate that the orientation of HAfp23 relative to the magnetic field can be changed, both in liquid crystalline d (GpG) and in axially stretched acrylamide gel (SAG), by simply altering the composition of the lipids in which the domain is embedded, i.e. by altering the charge distribution and/or shape of the protein:lipid aggregate. This approach is conceptually similar to previously reported methods which change a protein’s electrostatic surface by mutating exposed charged residues (Yao and Bax 2007) or that alter the shape of a multi-helical nucleic acid oligomer by altering the length of one of its helical stems (Zhang et al. 2006). However, our method only requires the production of a single protein preparation and instead changes the mixed micelle in which it is embedded.
Materials and Methods
Sample Preparation for NMR
Isotopically uniformly enriched 2H,13C,15N- or13C,15N-labeled HAfp23 (GLFGAIAGFI EGGWTGMIDG WYGSGKKKKD) was expressed as a fusion protein, preceded by the Igg binding domain B1 of streptococcal protein G. It was cleaved using factor Xa protease, and purified as described previously (Lorieau et al. 2010). The underlined sequence represents a polyionic "host" tail (Han and Tamm 2000) appended to aid in the preparation of the peptide sample. NMR samples contained 0.3–0.7 mM HAfp23 at pH 7.3 in 25 mM 2H-Tris buffer (Cambridge Isotopes), 7% 2H2O, and a formulation of isotropic bicelles (Vold et al. 1997) at a total lipid plus detergent concentration of 7–13% (w/w) for the SAG samples, and 3–6% for d (GpG) aligned samples. See below for details on sample alignment preparation, and see Table 1 for the composition of each sample used in the analysis. The lower lipid concentration for the d (GpG) samples was needed to preserve the liquid crystalline phase.
Table 1.
Saupe alignment tensor components for HAfp23 RDC datasets in different bicelle formulations aligned by stretched acrylamide gel and K-d (GpG) a,b
| Component/Tensor | A | B | C | D | E | F | G | H | I | |
|---|---|---|---|---|---|---|---|---|---|---|
| Szz | 11.0 | 17.6 | 11.4 | 13.3 | 12.0 | −5.3 | −11.8 | −12.7 | −12.1 | |
| −15.9 | −13.3 | −1.6 | −13.9 | −2.2 | 19.2 | 19.5 | 14.6 | 21.5 | ||
| 9.1 | 4.1 | 2.0 | 3.6 | 2.2 | −6.1 | −0.5 | 1.2 | −9.9 | ||
| −27.8 | −26.2 | −13.5 | −23.6 | −16.9 | 28.2 | 20.5 | 19.1 | 34.4 | ||
| 19.0 | 19.1 | 11.8 | 14.8 | 12.8 | −7.2 | −10.9 | −9.7 | −14.6 |
- Neutral bicelles in SAG: 0.6 mM 2H,13C,15N-labeled HAfp23, 25 mM2H-labeled Tris pH 7.3, 27 mM DMPC, 100 mM DOHPC, 16 mM DPC, 7% D2O in SAG (see Materials and Methods for details on the acrylamide gel formulation for this and other SAG samples).
- Bicelles + DMPS in SAG: 0.7 mM 2H,13C,15N-labeled HAfp23, 25mM2H-labeled Tris pH 7.3, 46 mM DMPC, 160 mM DOHPC, 8.1 mM DMPS, 10 mM DPC, 7% D2O in SAG.
- Bicelles + DMPS + DHPS in SAG: 0.7 mM 2H,13C,15N-labeled HAfp23, 25 mM2H-labeled Tris pH 7.3, 46 mM DMPC, 160 mM DOHPC, 8.1 mM DMPS, 46 mM DHPS, 10 mM DPC, 7% D2O in SAG.
- Bicelles + Et-DMPC in SAG: 0.6 mM 2H,13C,15N-labeled HAfp23, 25 mM2H-labeled Tris pH 7.4, 42 mM DMPC, 150 mM DOHPC, 10 mM Et-DMPC, 10 mM DPC, 7% D2O in SAG.
- Medium-sized bicelles in SAG: 0.7 mM 2H,13C,15N-labeled HAfp23, 25 mM 2H-labeled Tris pH 7.0, 73 mM DMPC, 144 mM DOHPC, 16 mM DPC, 7% D2O in SAG.
- Neutral bicelles in d (GpG): 0.3 mM 13C,15N-labeled HAfp23, 25 mM 2H-labeled Tris pH 7.3, 22 mM DMPC, 65 mM DOHPC, 10 mM DPC, 15 mg/ml d (GpG), 60 mM KCl, 7% D2O.
- Bicelles + DMPS in d (GpG): 0.3 mM 13C,15N-labeled HAfp23, 25 mM 2H-labeled Tris pH 7.4, 22 mM DMPC, 85 mM DOHPC, 6 mM DMPS, 10 mM DPC, 15 mg/ml d (GpG), 60 mM KCl, 7% D2O.
- Bicelles + DMPS + DHPS in d (GpG): 0.3 mM 13C,15N-labeled HAfp23, 25 mM 2H-labeled Tris pH 7.4, 22 mM DMPC, 85 mM DOHPC, 6 mM DMPS, 27 mM DHPS, 10 mM DPC, 15 mg/ml d (GpG), 60 mM KCl, 7% D2O.
- Bicelles + Et-DMPC in d (GpG): 0.3 mM 13C,15N-labeled HAfp23, 25 mM2H-labeled Tris pH 7.3, 25 mM DMPC, 82 mM DOHPC, 6 mM Et-DMPC, 10 mM DPC, 15 mg/ml d (GpG), 60 mM KCl, 7% D2O.
Tensor components are in units of Hz. Elements Sij of the 3×3 Cartesian alignment tensors were calculated from an SVD fit of the RDCs in Table S1 to the HAfp23 structure with PDB accession code 2KXA (Lorieau et al. 2010). They were then used to compute the appropriate 5-component representation of the alignment tensors shown here (Tolman 2002).
Isotropic bicelles were prepared by mixing neutral 1,2-di-O-hexyl-sn-glycero-3-phosphocholine (DOHPC; Avanti Lipids), with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; Avanti Lipids) at a molar ratio, q=[DOHPC]/[DMPC], of 0.33 ± 0.06, except for the medium-size bicelles, which had a q-ratio of 0.51. Charged lipids were added in powder form, without further purification, to NMR samples. 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS; Avanti Lipids) or 1,2-dihexanoyl-sn-glycero-3-phospho-L-serine (DHPS; Avanti Lipids) were used for negative charge doping, and 1,2-dimyristoyl-sn-glycero-3-ethylphosphocholine (Et-DMPC; Avanti Lipids) was used for adding positive charge. Charged lipids were added in a ~20–30% molar ratio relative to the DMPC concentration for the myristoylated charged lipids, or relative to the DOHPC concentration for DHPS, adding DOHPC or DMPC as needed to maintain the q-ratio of the bicelles. The q-ratios and absolute concentrations of lipids in the various NMR samples were quantified from one-dimensional 1H and 31P spectra by measuring the integrated intensity of the methyl and phosphate resonances.
Stretched Acrylamide Gel Samples
SAG samples were prepared for RDC measurement as described previously (Tycko et al. 2000; Meier et al. 2002; Lorieau et al. 2010), using neutral acrylamide (AA, 4.37% w/v), negatively charged 2-(acrylamido)-2-methyl-1-propanesulfonic acid (AMPS, 1.42% w/v), and 0.12% bis (acrylamide). The gel was cast in a cylindrical container of 5.4 mm diameter and then radially compressed to yield a final diameter of 4.2 mm inside the NMR sample tube (Chou et al. 2001).
Hydrogels were extensively dialyzed over multiple days, first in 50 ml of 25 mM Tris pH 7.4 buffer then in 50 ml of deionized water, before drying overnight at 37 °C. Dried hydrogel pellets were soaked into the bicelle-solubilized HAfp23 solutions for 3–4 days before transferring to the NMR sample tube for measurement. Separate SAG samples were prepared for bicelles with charged long-chain lipids. However, the higher diffusion rate of short-chain lipids, even in the presence of bicelles (Chou et al. 2004), made it possible to add a solution of detergent short-chain lipids directly to existing SAG NMR samples to produce a new alignment. These samples were given 3–4 days to equilibrate after addition of the concentrated (21% w/v) short-chain lipid solution. This equilibration time was adequate for complete mixing as judged by the 31P intensities of the sample and the protein RDCs, which did not change after more than a month of further equilibration. Alignment was also monitored by measuring the residual quadrupolar coupling (RQC) of the HDO deuterium signal. RQCs for SAG samples were between 0.8–1.2 Hz. NMR measurements were conducted at 32 °C.
Liquid Crystalline d (GpG) Samples
The d (GpG) liquid crystalline medium was prepared as previously described (Lorieau et al. 2008). The sodium salt of d (GpG) (Rasayan Inc, Encinitas, CA) was added directly to the sample to yield a ~15 mg/ml concentration, containing also 60 mM KCl. In an effective alternative protocol, not used for the data reported here, the d (GpG) is prepared by first dialyzing it in 20 mM K2HPO4 buffer at pH 7 using a 3.5 kDa molecular weight cutoff membrane, then lyophilizing it to a powder, prior to adding it to the sample. Dialysis lowers the total salt concentration by removing the lower-affinity sodium cation (Ghana et al. 1996). The pH of the sample was verified after addition of the d (GpG).
The concentration of d (GpG) was quantified by diluting the sample 1000-fold in 1 ml water and measuring the A260 after allowing the diluted sample to equilibrate for ~15–20 minutes, followed by vortexing. An extinction coefficient of 24.5 mg−1·cm−1·ml was used. Alignment was monitored by measuring the RQC of the HDO deuterium signal. RQCs for d (GpG) samples were between 13–18 Hz. The NMR experiments for d (GpG)-aligned samples were conducted at 27 °C.
Charged bicelle samples were prepared by adding charged lipids in dry powder form directly to d (GpG)-aligned neutral bicelle samples, followed by mixing in a thermomixer at 900 rpm and 37 °C, and subsequent vigorous vortexing.
NMR Experiments
1H-15N HSQC experiments were carried out at 500, 600, and 750 MHz nominal 1H Larmor frequencies, using Bruker Avance III spectrometers. The 750-MHz spectrometer was equipped with a room temperature TXI 750-MHz 1H/13C/15N/2H probe with three-axis gradients. The 600-MHz spectrometer was equipped with a cryogenically cooled QCI 600-MHz 1H/31P/13C/15N/2H probe, equipped with a z-axis gradient coil. The 500-MHz spectrometer was equipped with a room temperature QXI 500-MHz 1H/31P/13C/15N/2H probe with three-axis gradients.
The 1H-15N HSQC experiments used the Rance-Kay detection scheme (Kay et al. 1992). The 1H-15N RDCs were measured with an in-phase/anti-phase HSQC experiment with a Rance-Kay detection scheme (IPAP-RK) (Yao et al. 2009). Spectra were processed and analyzed using NMRPipe and Sparky software (Delaglio et al. 1995; Goddard and Kneller 2008).
Results
Bicelles are mixed bilayer-micelles composed of detergent and regular phospholipids, which self-assemble to form disc-shaped structures (Vold et al. 1997). Phospholipids that have a natural propensity for forming membrane bilayers in aqueous solution constitute the planar center of the disc, to produce a nearly ideal membrane mimetic (Vold et al. 1997), and the detergent partitions on the rim of the disc. Different detergents and phospholipids can be used to generate bicelles, with their molar ratio determining the size of the disc-shaped aggregate. As a starting point, we used bicelles composed of the detergent DOHPC, which is resistant to both acid- and base-catalyzed hydrolysis (Ottiger and Bax 1999), and the long-chain phospholipid DMPC. At detergent:phospholipid molar ratios, q <~0.7, bicelles are relatively small, with a mass ranging from ca 22 kDa for q=0.15, to more than 80 kDa for q=0.7, and diameters for the planar component of the bicelle ranging from <20 to more than 40 Å (Chou et al. 2004). The shape of small bicelles resembles that of an oblate spheroid, which tumbles nearly isotropically (Cantor and Schimmel 1980). The relatively small size of the bicelles results in rotational correlation times that are sufficiently short to produce high-resolution NMR spectra of membrane proteins embedded within them. This contrasts with liquid crystalline aligned bicelles, obtained when using q > ~3, which adopt a ‘swiss cheese’-like bilayer morphology (Gaemers and Bax 2001). Molecules anchored in such strongly oriented bicelles become highly ordered relative to the magnetic field and require solid-state NMR technology to study them (Sanders and Schwonek 1992; De Angelis and Opella 2007).
Small bicelles can be aligned very weakly relative to the magnetic field by embedding them in a stretched acrylamide hydrogel (Chou et al. 2002) or by using a detergent-compatible liquid crystalline suspension (Douglas et al. 2007; Lorieau et al. 2008). We demonstrate that the orientation of the HAfp23-bicelle complex relative to the magnetic field can be altered by changing the charge of the bicelle lipids or detergent, both in liquid crystalline d (GpG) and in stretched acrylamide gel, doped with 2-acrylamido-2-methyl-1-propanesulfonic acid (AMPS) (Cierpicki and Bushweller 2004).
As mentioned above, the size of the bicelle can be adjusted by changing the molar ratio, q, of long-chain lipids to the detergent (Vold and Prosser 1996). The charge of a bicelle is easily changed by replacing a fraction (typically ~20–30%) of the detergent molecules by acidic or basic short-chain lipids, or by replacing a similar fraction of the DMPC component by charged long-chain phospholipids (Struppe et al. 2000). For negative charge doping, we used DHPS, to substitute for neutral DOHPC, while negative and positive doping of the bilayer part of the bicelle was accomplished by substituting DMPS or Et-DMPC, respectively, for neutral DMPC. The association of the charged DMPS and DHPS lipids to the bicelle was confirmed by translational diffusion measurements (Supplementary Materials), with the DMPS remaining tightly associated at all measured lipid concentrations and the DHPS also significantly partitioning on the bicelles at total lipid concentrations (lipid plus detergent) above 5–7% (w/w).
Five different bicelle-HAfp23 samples were prepared: four with q-ratios in the 0.33 ± 0.06 range, resulting in small bicelles, and one with a q-ratio of 0.51, yielding a medium-sized bicelle. The calculated diameter of the planar region of the bicelle (Vold and Prosser 1996) corresponds to ca 50 Å for q=0.33 and ca 80 Å for q=0.51. This compares with a bilayer thickness of ca 40 Å. Including the diameter of the hemi-toroidal ring surrounding the idealized bicelle, which adds 40 Å to its diameter, this corresponds to aspect ratios of ~2.2 and ~3 for the small and medium sized bicelles, respectively. However, translational and rotational diffusion rates (Chou et al. 2004; Lorieau et al. 2011) indicate that in practice the bicelles are substantially smaller than calculated for the idealized model, with presumably smaller aspect ratios.
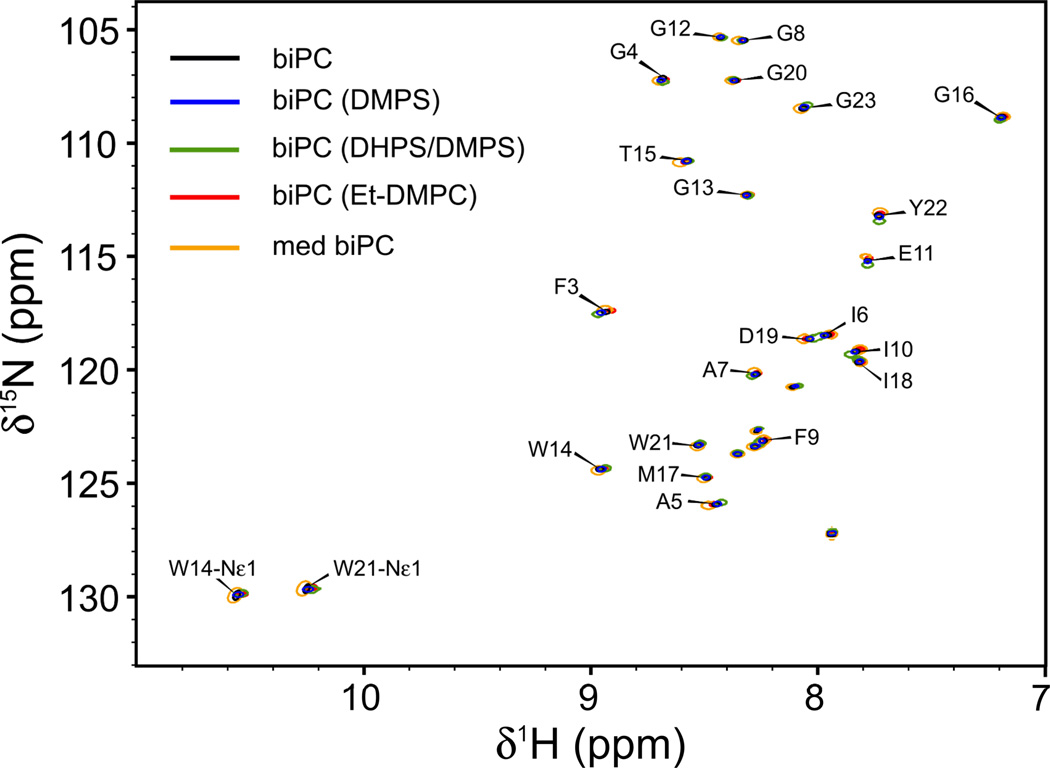
For collecting orthogonal RDC datasets, it is prerequisite that the structure of the protein is not significantly impacted by the composition of the bicelles, a condition that is easily validated from its 1H-15N-HSQC spectrum, which is exquisitely sensitive to even very minor structural perturbations. As can be seen from the 1H-15N HSQC spectra recorded for HAfp23, the spectra recorded in the presence of the five different bicelle types perfectly superimpose (Figure 1). The line widths obtained for the four small-bicelle samples (q=~0.33) are very similar, confirming that the rotational tumbling times for these systems are comparable. The 1H and 15N line widths increase for HAfp23 in the larger bicelles (q=0.51), as these bicelles have a rotational correlation time that is ca. 1.8 times greater than for the small bicelles (Lorieau et al. 2011).
Fig. 1.
Superimposed 1H-15N HSQC spectra of the HAfp23 peptide in isotropic bicelles (biPC) of different size and charge at pH 7. Shown are spectra for small bicelles (biPC, q=0.33 ± 0.06) without net charge (black), bicelles with DMPS (blue), DMPS/DHPS (green), Et-DMPC (red), and medium-sized neutral bicelles (med biPC, q=0.51, orange). Unlabeled cross peaks correspond to the SGKKKKD polyionic "host" tail (Han and Tamm 2000), used to aid in the sample preparation (Lorieau et al. 2010). Spectra were collected in the absence of alignment media.
The Saupe tensor defines the linear relationship between RDCs and the directional cosines of the vectors between nuclei (Losonczi et al. 1999).
| [1] |
where ϕp qrepresents the angle between internuclear vector p and axis q of the molecular coordinate frame; and Syy, Szz, Sxy, Sxz, and Syz are the five independent elements of the traceless, symmetric Saupe matrix that correlate the internuclear vector orientations to the corresponding observed dipolar coupling values, D1,..,n. The alignment of each sample is defined by its own Saupe matrix elements (Table 1). Diagonalization of the Saupe matrix yields the commonly used Da, and Rh parameters, defining the strength and rhombicity of the alignment (Bax 2003).
The orthogonality between RDC datasets can be assessed by calculating the normalized scalar product of the corresponding Saupe matrices (Sass et al. 1999). The absolute value for the alignment tensor dot products of the different samples range in value from 0.74 to 0.997 (Table 2), indicating that there are no two datasets that are mutually fully orthogonal. Indeed, the alignment tensors of the q=0.51 neutral bicelles and the negatively charged (DMPS+DHPS) smaller (q=0.33) bicelles in SAG medium are nearly indistinguishable. No clear trends emerge in the physical properties that govern the orientation of the alignment frame for the different datasets, though contributions from both electrostatic and steric forces clearly play a significant role.
Table 2.
Normalized scalar products of the molecular alignment tensors of HAfp23 aligned in small bicelles of different charge and shape.a
| A | B | C | D | E | F | G | H | |
|---|---|---|---|---|---|---|---|---|
| A: neutral bicelles in SAG | ||||||||
| B: bicelle + DMPS in SAG | 0.976 | |||||||
| C: bicelle + DMPS + DHPS in SAG | 0.901 | 0.959 | ||||||
| D: bicelle + Et-DMPC in SAG | 0.985 | 0.995 | 0.926 | |||||
| E: medium bicelles in SAG | 0.919 | 0.967 | 0.997 | 0.941 | ||||
| F: bicelles in d (GpG) | −0.939 | −0.886 | −0.743 | −0.928 | −0.784 | |||
| G: bicelles + DMPS in d (GpG) | −0.941 | −0.947 | −0.822 | −0.970 | −0.840 | 0.941 | ||
| H: bicelles + DMPS + DHPS in d (GpG) | −0.934 | −0.964 | −0.870 | −0.977 | −0.886 | 0.917 | 0.991 | |
| I: bicelles + Et-DMPC in d (GpG) | −0.984 | −0.951 | −0.845 | −0.976 | −0.873 | 0.982 | 0.959 | 0.946 |
See Table 1 for a listing of sample compositions.
When dealing with more than two alignment tensors, a convenient and effective method for decomposing these into orthogonal sets relies on a singular value decomposition (SVD). SVD analysis of different alignment conditions has previously been carried out directly on the experimental RDCs to yield orthogonalized linear combinations (OLC) of RDC datasets without requiring a molecular structure (Ruan and Tolman 2005; Ruan et al. 2008). However, while generating linearly independent RDC data is convenient and can be carried out in the absence of a reference structure, the corresponding Saupe matrices can deviate significantly from being orthogonal. In particular, for HAfp23 which contains a highly non-uniform distribution of its 15N-1H bond vectors, the orthogonalized linear combinations of RDCs correspond to Saupe matrices with normalized scalar products as large as 0.62, and even higher for the weakest OLCs (Table S11).
Instead, we therefore carried out the SVD analysis directly on the alignment tensors (Table 1) generated from the experimental RDCs and the coordinates of the peptide (PDB entry 2KXA). The alignment tensors were represented by 5D vectors of the form (Tolman 2002). The numerical coefficients are required to ensure invariance of dot products under rotations of the coordinate frame, the behavior exhibited by Cartesian alignment tensors in matrix form. Values of normalized dot products calculated using this representation match the previously introduced normalized scalar products (Sass et al. 1999).
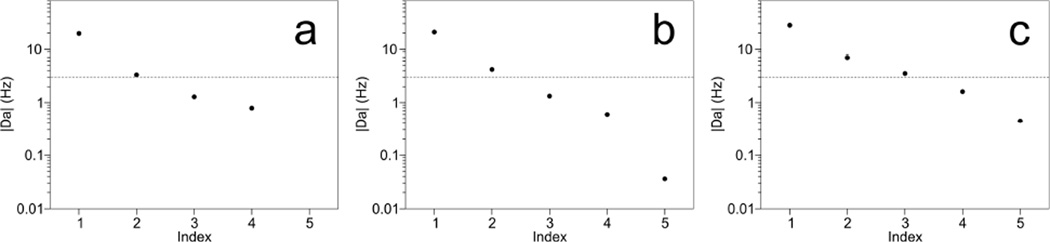
A new set of five orthogonal alignment tensors was generated from the Saupe matrices obtained for the different samples (Table 3), after first normalizing the tensors of Table 1 to an alignment strength, Da, of 10 Hz. The normalization of the alignment tensors, which have fitted Da values that range in absolute value between 10 and 22 Hz, ensures that each RDC dataset contributes about equally to the final orthogonalized tensor set. The number of usable orthogonal tensors contained in the experimental RDC datasets then can be estimated by the number of orthogonalized tensors that have Da values far above the experimental RDC measurement error of ca 0.5 Hz. The results show that in SAG or d (GpG) medium, the decomposition yields only two orthogonal alignment tensors with amplitudes that are far above the measurement error (Figure 2a,b). However, when combining the RDCs measured in SAG and d (GpG) for the five different types of bicelles, three orthogonal alignment orientations have magnitudes that are well above the measurement error. Refinement of the originally deposited structure, with the three additional orthogonalized sets of RDCs used as input restraints, resulted in a structure that differed by 0.28 Å from the PDB structure (entry 2KXA), and interestingly showed a decrease in Q-factor from 31 to 21% for the fourth orthogonal set of (very small) RDCs which were not used during refinement because of their considerably lower experimental precision.
Table 3.
Saupe alignment tensor components obtained after orthogonalization of the normalized alignment tensors from Table 1.a
| Component/Tensor | Tensor 1 | Tensor 2 | Tensor 3 | Tensor 4 | Tensor 5 | |
|---|---|---|---|---|---|---|
| Szz | −24.31 | −6.22 | 3.71 | 0.33 | −0.59 | |
| 24.41 | −10.56 | −2.54 | 1.27 | 0.07 | ||
| −6.85 | −0.58 | −5.11 | −1.17 | −0.58 | ||
| 44.17 | −1.36 | 2.43 | −1.95 | −0.18 | ||
| −26.44 | −6.45 | −0.38 | −2.09 | 0.45 | ||
| Da | −28.13 | −6.92 | 3.52 | 1.61 | −0.45 | |
| Rh | 0.55 | 0.13 | 0.27 | 0.33 | 0.44 |
Tensor components are in units of Hz and have been generated with a SVD of the tensors from Table 1, after scaling these to a Da of 10 Hz.
Fig. 2.
Da values obtained after best-fitted alignment tensors were decomposed into orthogonal sets for (a) d (GpG), (b) stretched acrylamide gel, and (c) the combination of both. The input tensors were normalized to a Da value of 10 Hz. The dashed line at 3 Hz marks a 6:1 ratio for the magnitude of the alignment tensor over the ~0.5 Hz experimental uncertainty in the RDCs. For the combined sets of RDCs measured in d (GpG) and SAG media (c), the reconstructed sets of experimental RDCs that correspond to these orthogonalized Saupe matrices (Table 3) fit to the deposited coordinates (PDB entry 2KXA) with Q-factors of 13, 26, 10, 31 and 94%. A further structural refinement which includes the first three sets of reconstructed RDCs (using force constants that scale linearly with Da) resulted in a structure with Q-factors of 4%, 12% and 9%, and a backbone heavy-atom RMSD of 0.28Å for residues 3–22 from PDB structure 2KXA. The fourth component, which was not used in the refinement, showed an improved Q-factor of 21%. The Q-factor of the fifth orthogonalized set is completely dominated by experimental uncertainties in the RDC measurements.
Concluding remarks
Our results demonstrate that sufficiently unique RDC datasets can be obtained for a peptide or protein embedded in a membrane-mimicking bicelle by simply varying its charge and shape. For the small HAfp23 domain, the close agreement between the additional independent RDC data and its previously determined structure simply confirmed the accuracy of these coordinates. For more complex structures, however, availability of multiple alignment tensors can resolve the orientational degeneracy between different fragments of the protein (Al-Hashimi et al. 2000). We anticipate this will be particularly useful in systems that include both a transmembrane and cytosolic domain, where accurate determination of the relative orientation and dynamics of these components can remain challenging, even in the presence of a single set of RDC data. Addition of detergent to the protein and bicelle containing NMR sample is particularly simple, as the powder detergent can be added to the sample after a first set of RDCs has been measured, either by simple mixing for the d (GpG) sample, or by resoaking the gel in a small volume of detergent containing buffer.
Supplementary Material
ACKNOWLEDGMENT
We thank Annie Aniana for help with protein expression and purification, Nicolas A. Bax for measuring the cmc of DHPS, and Dennis A. Torchia for discussions and comments. This work was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH) and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
Footnotes
Electronic supplementary material. The online version of this article (doi:xxx) contains supplementary material, which is available to authorized users.
References
- Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK, Prestegard JH. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J. Magn. Reson. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Meth. Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- Bertini I, Del Bianco C, Gelis I, Katsaros N, Luchinat C, Parigi G, Peana M, Provenzani A, Zoroddu MA. Experimentally exploring the conformational space sampled by domain reorientation in calmodulin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6841–6846. doi: 10.1073/pnas.0308641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Kursula P, Luchinat C, Parigi G, Vahokoski J, Wilmanns M, Yuan J. Accurate Solution Structures of Proteins from X-ray Data and a Minimal Set of NMR Data: Calmodulin-Peptide Complexes As Examples. J. Am. Chem. Soc. 2009;131:5134–5144. doi: 10.1021/ja8080764. [DOI] [PubMed] [Google Scholar]
- Blackledge M. Recent progress in the study of biomolecular structure and dynamics in solution from residual dipolar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2005;46:23–61. [Google Scholar]
- Briggman KB, Tolman JR. De Novo determination of bond orientations and order parameters from residual dipolar couplings with high accuracy. J. Am. Chem. Soc. 2003;125:10164–10165. doi: 10.1021/ja035904+. [DOI] [PubMed] [Google Scholar]
- Cantor CR, Schimmel PR. Biophysical Chemistry. San Francisco: Freeman; 1980. [Google Scholar]
- Chou JJ, Baber JL, Bax A. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in a stretched polyacrylamide gel. J. Am. Chem. Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J. Am. Chem. Soc. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- Clore GM, Garrett DS. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 1999;121:9008–9012. [Google Scholar]
- Clore GM, Starich MR, Bewley CA, Cai ML, Kuszewski J. Impact of residual dipolar couplings on the accuracy of NMR structures determined from a minimal number of NOE restraints. J. Am. Chem. Soc. 1999;121:6513–6514. [Google Scholar]
- Clore GM, Starich MR, Gronenborn AM. Measurement of Residual Dipolar Couplings of Marcomolecules Aligned in the Nematic Phase of a Colloidal Suspension of Rod-Shaped Viruses. J. Am. Chem. Soc. 1998;120:10571–10572. [Google Scholar]
- Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 1998;120:6836–6837. [Google Scholar]
- De Angelis AA, Opella SJ. Bicelle samples for solid-state NMR of membrane proteins. Nat. Protoc. 2007;2:2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe - a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaemers S, Bax A. Morphology of three lyotropic liquid crystalline biological NMR media studied by translational diffusion anisotropy. J. Am. Chem. Soc. 2001;123:12343–12352. doi: 10.1021/ja011967l. [DOI] [PubMed] [Google Scholar]
- Ghana R, Walss C, Walmsley JA. Sodium and potassium ion-promoted formation of supramolecular aggregates of 2'-deoxyguanylyl-(3'–5')-2'-deoxyguanosine. J. Biomol. Struct. Dyn. 1996;14:101–110. doi: 10.1080/07391102.1996.10508933. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 2008. [Google Scholar]
- Han X, Tamm LK. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- Hus J-C, Salmon L, Bouvignies G, Lotze J, Blackledge M, Brueschweiler R. 16-Fold Degeneracy of Peptide Plane Orientations from Residual Dipolar Couplings: Analytical Treatment and Implications for Protein Structure Determination. J. Am. Chem. Soc. 2008;130:15927–15937. doi: 10.1021/ja804274s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hus JC, Peti W, Griesinger C, Bruschweiler R. Self-consistency analysis of dipolar couplings in multiple alignments of ubiquitin. J. Am. Chem. Soc. 2003;125:5596–5597. doi: 10.1021/ja029719s. [DOI] [PubMed] [Google Scholar]
- Kamen DE, Cahill SM, Girvin ME. Multiple alignment of membrane proteins for measuring residual dipolar couplings using lanthanide ions bound to a small metal chelator. J. Am. Chem. Soc. 2007;129:1846–1847. doi: 10.1021/ja067089e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Keifer P, Saarinen T. Pure Absorption Gradient Enhanced Heteronuclear Single Quantum Correlation Spectroscopy with Improved Sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- Lorieau J, Yao LS, Bax A. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J. Am. Chem. Soc. 2008;130:7536–7537. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, Louis JM, Bax A. Whole-Body Rocking Motion of a Fusion Peptide in Lipid Bilayers from Size-Dispersed (15) N NMR Relaxation. J. Am. Chem. Soc. 2011;133:14184–14187. doi: 10.1021/ja2045309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J. Magn. Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- Meier S, Haussinger D, Grzesiek S. Charged acrylamide copolymer gels as media for weak alignment. J. Biomol. NMR. 2002;24:351–356. doi: 10.1023/a:1021609207024. [DOI] [PubMed] [Google Scholar]
- Meiler J, Prompers JJ, Peti W, Griesinger C, Bruschweiler R. Model-free approach to the dynamic interpretation of residual dipolar couplings in globular proteins. J. Am. Chem. Soc. 2001;123:6098–6107. doi: 10.1021/ja010002z. [DOI] [PubMed] [Google Scholar]
- Mueller GA, Choy WY, Yang DW, Forman-Kay JD, Venters RA, Kay LE. Global folds of proteins with low densities of NOEs using residual dipolar couplings: Application to the 370-residue maltodextrin-binding protein. J. Mol. Biol. 2000;300:197–212. doi: 10.1006/jmbi.2000.3842. [DOI] [PubMed] [Google Scholar]
- Ottiger M, Bax A. Bicelle-based liquid crystals for NMR-measurement of dipolar couplings at acidic and basic pH values. J. Biomol. NMR. 1999;13:187–191. doi: 10.1023/a:1008395916985. [DOI] [PubMed] [Google Scholar]
- Peti W, Meiler J, Bruschweiler R, Griesinger C. Model-free analysis of protein backbone motion from residual dipolar couplings. J. Am. Chem. Soc. 2002;124:5822–5833. doi: 10.1021/ja011883c. [DOI] [PubMed] [Google Scholar]
- Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- Ramirez BE, Bax A. Modulation of the alignment tensor of macromolecules dissolved in a dilute liquid crystalline medium. J. Am. Chem. Soc. 1998;120:9106–9107. [Google Scholar]
- Rodriguez-Castaneda F, Haberz P, Leonov A, Griesinger C. Paramagnetic tagging of diamagnetic proteins for solution NMR. Magn. Reson. Chem. 2006;44:S10–S16. doi: 10.1002/mrc.1811. [DOI] [PubMed] [Google Scholar]
- Ruan K, Briggman KB, Tolman JR. De novo determination of internuclear vector orientations from residual dipolar couplings measured in three independent alignment media. J. Biomol. NMR. 2008;41:61–76. doi: 10.1007/s10858-008-9240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan K, Tolman JR. Composite alignment media for the measurement of independent sets of NMR residual dipolar couplings. J. Am. Chem. Soc. 2005;127:15032–15033. doi: 10.1021/ja055520e. [DOI] [PubMed] [Google Scholar]
- Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- Sanders CR, Schwonek JP. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry. 1992;31:8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- Sass H-J, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- Sass J, Cordier F, Hoffmann A, Rogowski M, Cousin A, Omichinski JG, Lowen H, Grzesiek S. Purple membrane induced alignment of biological macromolecules in the magnetic field. J. Am. Chem. Soc. 1999;121:2047–2055. [Google Scholar]
- Saupe A, Englert G. High-resolution nuclear magnetic resonance spectra of oriented molecules. Phys. Rev. Lett. 1963;11:462–464. [Google Scholar]
- Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- Struppe J, Whiles JA, Vold RR. Acidic phospholipid bicelles: A versatile model membrane system. Biophys. J. 2000;78:281–289. doi: 10.1016/S0006-3495(00)76591-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XC, Otting G. Paramagnetic labelling of proteins and oligonucleotides for NMR. J. Biomol. NMR. 2010;46:101–112. doi: 10.1007/s10858-009-9331-1. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- Tolman JR. A novel approach to the retrieval of structural and dynamic information from residual dipolar couplings using several oriented media in biomolecular NMR spectroscopy. J. Am. Chem. Soc. 2002;124:12020–12030. doi: 10.1021/ja0261123. [DOI] [PubMed] [Google Scholar]
- Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear Magnetic Dipole Interactions in Field-Oriented Proteins - Information For Structure Determination in Solution. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem. Rev. 2006;106:1720–1736. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- Tycko R, Blanco FJ, Ishii Y. Alignment of biopolymers in strained gels: A new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J. Am. Chem. Soc. 2000;122:9340–9341. [Google Scholar]
- Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J. Am. Chem. Soc. 2003;125:9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- Vold RR, Prosser RS. Magnetically oriented phospholipid bilayered micelles for structural studies of polypeptides. Does the ideal bicelle exist? Journal of Magnetic Resonance Series B. 1996;113:267–271. [Google Scholar]
- Vold RR, Prosser RS, Deese AJ. Isotropic solutions of phospholipid bicelles: A new membrane mimetic for high-resolution NMR studies of polypeptides. J. Biomol. NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- Wohnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J. Am. Chem. Soc. 2003;125:13338–13339. doi: 10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]
- Yao L, Vogeli B, Torchia DA, Bax A. Simultaneous NMR study of protein structure and dynamics using conservative mutagenesis. J. Phys. Chem. B. 2008;112:6045–6056. doi: 10.1021/jp0772124. [DOI] [PubMed] [Google Scholar]
- Yao LS, Bax A. Modulating protein alignment in a liquid-crystalline medium through conservative mutagenesis. J. Am. Chem. Soc. 2007;129:11326–11327. doi: 10.1021/ja073937+. [DOI] [PubMed] [Google Scholar]
- Yao LS, Ying JF, Bax A. Improved accuracy of N-15-H-1 scalar and residual dipolar couplings from gradient-enhanced IPAP-HSQC experiments on protonated proteins. J. Biomol. NMR. 2009;43:161–170. doi: 10.1007/s10858-009-9299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Sun XY, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- Zidek L, Padrta P, Chmelik J, Sklenar V. Internal consistency of NMR data obtained in partially aligned biomacromolecules. J. Magn. Reson. 2003;162:385–395. doi: 10.1016/s1090-7807(03)00116-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.