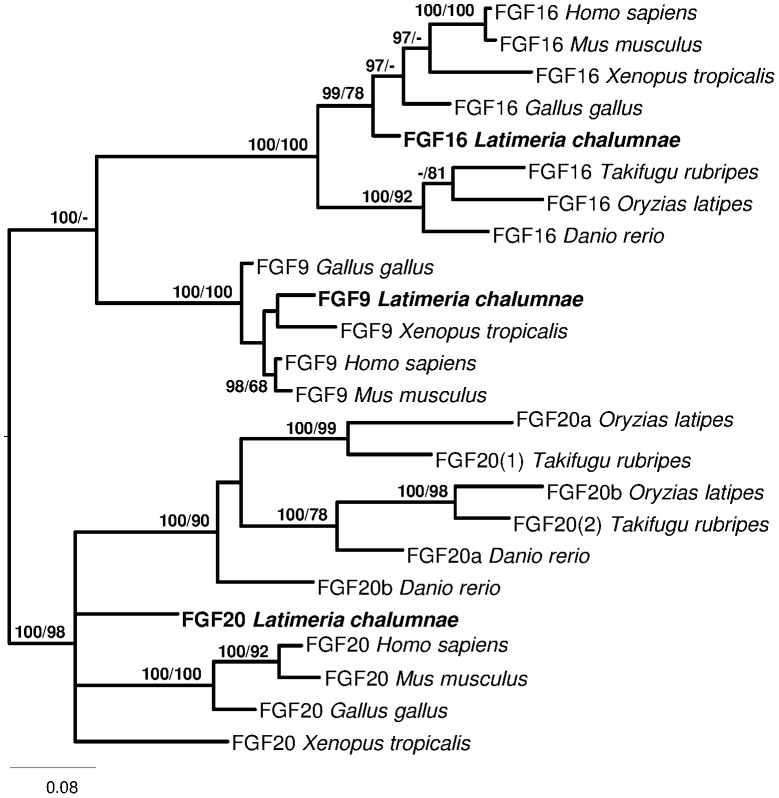
Figure 6. Phylogenetic tree of FGF9, FGF16, and FGF20.
Phylogenetic analysis of amino acid sequences of the vertebrate FGF9/16/20. Midpoint rooting. Total characters: 237, constant: 91, parsimony non-informative: 35, parsimony informative: 111. Numbers close to nodes represent posterior probability in Bayesian Inference/bootstrap percentage in Maximum Parsimony. Danio rerio (FGF16: ENSDART00000061928; FGF20a: NP_001032180.1; FGF20b: NP_001034261.1); Gallus gallus (FGF9: NP_989730.1; FGF16; NP_001038115.1; FGF20: XP_426335.2); Homo sapiens (FGF9: NP_002001.1; FGF16: NP_003859.1; FGF20: NP_062825.1); Latimeria chalumnae (FGF9: manually inferred from JH128123; FGF16: ENSLACT00000011509; FGF20: ENSLACT00000014939); Mus musculus (FGF9: ADL60500.1; FGF16: BAB16405.1; FGF20: NP_085113.2); Oryzias latipes (FGF16: ENSORLT00000007651; FGF20a: ENSORLT00000012578; FGF20b: ENSORLT00000025767); Takifugu rubripes (FGF16: ENSTRUT00000021181; FGF20(1): ENSTRUT00000008788; FGF20(2): ENSTRUT00000039390); Xenopus tropicalis (FGF9: XP_002938621.1; FGF16: ENSXETT00000009790; FGF20: NP_001137399.1). Latimeria menadoensis is missing in this analysis because FGF9 and FGF20 are poorly or not expressed in the transcriptomes.