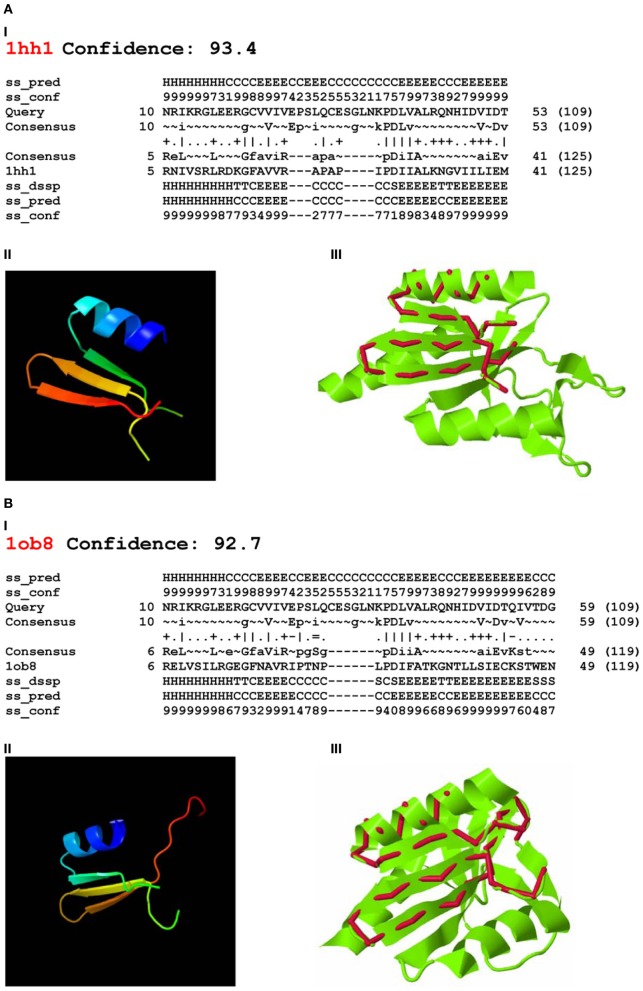
Figure 3.
Phyre-2 comparisons of the endonuclease domain of the Drosophila melanogaster R2 non-LTR retrotransposon with HHpred Hits. Hits: (A) Holliday-junction resolving enzyme of Sulfolobus solfataricus (1hh1); (B) Holliday-junction resolvase of S. solfataricus (1ob8). I—secondary structure prediction, abbreviations as in Figure 2; II—3D structure of the queried sequence based on HHpred alignment; III—comparison of 3D structures. Red, queried sequence; green, 3D structure (PDB). Confidence, probability (from 0 to 100) that a match between the query sequence and a given template is a true homology, with >90% confidence that the query protein adopts the overall fold shown and the core is modeled at high accuracy (2–4 Å rmsd from native, true structure).