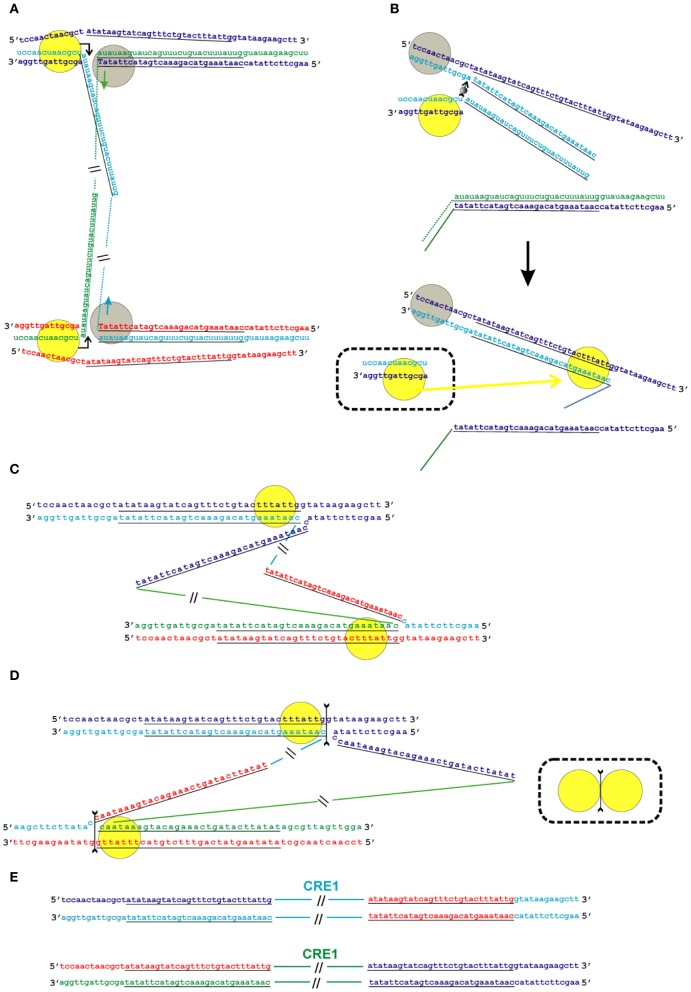
Figure 6.
Model of the transposition of the R2 group of non-LTR retrotransposons for which the endonuclease makes the second DNA nick downstream of the first one. Sequences correspond to target sites of the Crithidia fasciculata CRE1 non-LTR retrotransposon localized within a miniexon. The sequences that correspond to the duplications of the target site are underlined. A schematic presentation of two retrotransposon proteins bound to the RNA co-transcript that contains both the retrotransposon sequence and sequences that flank the target site is not shown. (A) Proteins, while bound to the RNAs, bind DNA target sites that are located on the homologous chromosomes. The proteins contribute to the melting of the DNA strands, and RNA/DNA duplexes (R-loops) are formed. Dotted lines correspond to the sequences of the mobile element RNAs. The protein bound close to the 5′-end of the RNA (yellow circle) makes the first ssDNA nick (depicted by a bent arrow). Target primed reverse transcription is started by the protein bound close to the 3′-end of the RNA (gray circles). The nucleotides that are donors of the hydroxyl group are depicted in upper-case letters. (B) Here, only events that occur within a single chromosome are shown. When synthesis of the retrotransposon cDNA (solid line) is complete, the protein (gray circles) rests against the RNA/DNA hybrid. Subsequently, this protein jumps to the corresponding free single DNA strand (the jump is depicted by an arrow with a small gray circle) and continues synthesis of the complementary DNA strand. The protein marked by a yellow circle moves (yellow arrow) from the end of the RNA/DNA hybrid to the newly synthesized dsDNA. It is most likely that the RNA in the RNA/DNA hybrids is digested by endogenous host RNase H. The bold black arrow shows the change in conformation of the DNA strands at the 5′-end of the target sites. The sequence that corresponds to the target site duplication anneals to the complementary strand and the protein (yellow circle) moves (yellow arrow) to the end of the dsDNA. (C) As a result of the processes described above, a typical Holliday junction structure is formed, with two proteins (yellow circles) bound to the target sites that are located in the homologous chromosomes. (D) Owing to the spatial complexity of the structures shown in the figure, two proteins (yellow circles) are shown separately; however, according to the model, they form a dimer that shows Holliday junction-resolving activity. The second cut is shown by an arrow with two feathers. (E) The copies of the retrotransposon that are integrated into the homologous chromosomes are shown. The host DNA polymerase completes the synthesis of the second DNA strand that corresponds to the mobile element.