Abstract
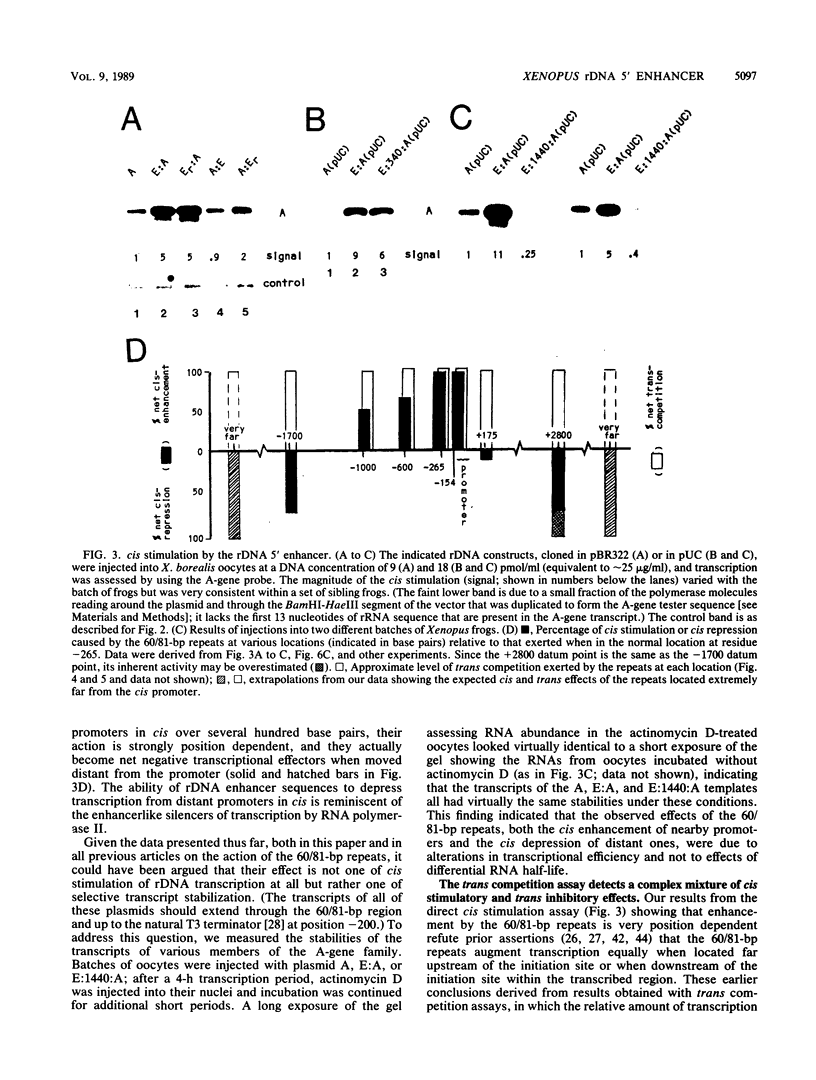
Although it is generally believed that the 60- and 81-base-pair (60/81-bp) repeats of the Xenopus laevis ribosomal DNA (rDNA) spacer are position-independent transcriptional enhancers, this has not been shown directly. We have now developed a critical assay which proves that the 60/81-bp repeats do, in fact, stimulate transcription from promoters in cis and that they function in both orientations and when up to 1 kilobase pair from the initiation site. However, contrary to the widely accepted view, these elements are found to be highly position dependent, for they have no net effect when downstream of the initiation site within the transcribed region and they behave as transcriptional silencers of promoters in cis when moved greater than 2 kilobase pairs upstream of the initiation site. The 60/81-bp elements therefore are position-dependent 5' enhancers. We also found that this rDNA enhancer was polymerase I specific and that it was composed of duplicated, individually functional elements. Finally, we report an in vitro system that reproduces both cis enhancement and trans competition by the 60/81-bp repeats. Sequential-addition studies in this system demonstrated that the rDNA enhancer functions in trans at or before establishment of the stable transcription complex, not subsequently at each round of transcription.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Mangin M., Weiner A. M. Orientation-dependent transcriptional activator upstream of a human U2 snRNA gene. Mol Cell Biol. 1985 Jul;5(7):1560–1570. doi: 10.1128/mcb.5.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark C., Weller P., Zabielski J., Janson L., Pettersson U. A distant enhancer element is required for polymerase III transcription of a U6 RNA gene. Nature. 1987 Jul 23;328(6128):356–359. doi: 10.1038/328356a0. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Brady J., Loeken M. R., Khoury G. Interaction between two transcriptional control sequences required for tumor-antigen-mediated simian virus 40 late gene expression. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7299–7303. doi: 10.1073/pnas.82.21.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S. J., Reeder R. H. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell. 1983 Oct;34(3):989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Palla F., Tebb G., Mattaj I. W., Philipson L. Properties of a U1 RNA enhancer-like sequence. Nucleic Acids Res. 1987 Mar 25;15(6):2403–2416. doi: 10.1093/nar/15.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Plon S. E., Wang J. C. The use of psoralen-modified DNA to probe the mechanism of enhancer action. Cell. 1986 May 23;45(4):567–574. doi: 10.1016/0092-8674(86)90288-6. [DOI] [PubMed] [Google Scholar]
- Culotta V. C., Wilkinson J. K., Sollner-Webb B. Mouse and frog violate the paradigm of species-specific transcription of ribosomal RNA genes. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7498–7502. doi: 10.1073/pnas.84.21.7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. A complex array of sequences enhances ribosomal transcription in Xenopus laevis. J Mol Biol. 1987 Aug 20;196(4):813–827. doi: 10.1016/0022-2836(87)90407-4. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell. 1986 Jan 31;44(2):313–318. doi: 10.1016/0092-8674(86)90765-8. [DOI] [PubMed] [Google Scholar]
- Evans T., Reitman M., Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Green J., Brady J., Khoury G. 72-bp element contains a critical control region for SV40 late expression in Xenopus laevis oocytes. Virology. 1987 Aug;159(2):339–349. doi: 10.1016/0042-6822(87)90472-7. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Horowitz H., Platt T. Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res. 1982 Sep 25;10(18):5447–5465. doi: 10.1093/nar/10.18.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Xenopus ribosomal gene enhancers function when inserted inside the gene they enhance. Nucleic Acids Res. 1985 Dec 20;13(24):8999–9009. doi: 10.1093/nar/13.24.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. RNA polymerase specificity of mRNA production and enhancer action. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6677–6681. doi: 10.1073/pnas.83.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Havercroft J. C., Chow L. T., Cleveland D. W. Four unique genes required for beta tubulin expression in vertebrates. Cell. 1983 Mar;32(3):713–724. doi: 10.1016/0092-8674(83)90057-0. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Sollner-Webb B., Cleveland D. W. Surprising S1-resistant trimolecular hybrids: potential complication in interpretation of S1 mapping analyses. Mol Cell Biol. 1985 Oct;5(10):2842–2846. doi: 10.1128/mcb.5.10.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Miwa T., Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory roles in expression of the human alpha-cardiac actin gene. Mol Cell Biol. 1987 Aug;7(8):2803–2813. doi: 10.1128/mcb.7.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Eaton S., Calame K. Purified mu EBP-E binds to immunoglobulin enhancers and promoters. Mol Cell Biol. 1988 Nov;8(11):4972–4980. doi: 10.1128/mcb.8.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Schaffner W. Correct transcription of a cloned mouse immunoglobulin gene in vivo. Proc Natl Acad Sci U S A. 1983 Jan;80(2):417–421. doi: 10.1073/pnas.80.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Viral and cellular transcription enhancers. Oxf Surv Eukaryot Genes. 1985;2:24–48. [PubMed] [Google Scholar]
- Pikaard C. S., Reeder R. H. Sequence elements essential for function of the Xenopus laevis ribosomal DNA enhancers. Mol Cell Biol. 1988 Oct;8(10):4282–4288. doi: 10.1128/mcb.8.10.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Pennock D., McStay B., Roan J., Tolentino E., Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987 Sep 25;15(18):7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G. The mechanism of nucleolar dominance in Xenopus hybrids. Cell. 1984 Aug;38(1):38–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Wildeman A., Chambon P. A trans-acting factor is responsible for the simian virus 40 enhancer activity in vitro. Nature. 1985 Feb 7;313(6002):458–463. doi: 10.1038/313458a0. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., McKnight S. L. Accurate transcription of cloned Xenopus rRNA genes by RNA polymerase I: demonstration by S1 nuclease mapping. Nucleic Acids Res. 1982 Jun 11;10(11):3391–3405. doi: 10.1093/nar/10.11.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Wilkinson J. A., Roan J., Reeder R. H. Nested control regions promote Xenopus ribosomal RNA synthesis by RNA polymerase I. Cell. 1983 Nov;35(1):199–206. doi: 10.1016/0092-8674(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Spinelli G., Ciliberto G. Functional activity and chromatin configuration of SV40 enhancer injected in Xenopus laevis oocytes. Nucleic Acids Res. 1985 Nov 25;13(22):8065–8081. doi: 10.1093/nar/13.22.8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Tower J., Culotta V. C., Sollner-Webb B. Factors and nucleotide sequences that direct ribosomal DNA transcription and their relationship to the stable transcription complex. Mol Cell Biol. 1986 Oct;6(10):3451–3462. doi: 10.1128/mcb.6.10.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., Calame K. SV40 enhancer-binding factors are required at the establishment but not the maintenance step of enhancer-dependent transcriptional activation. Cell. 1986 Oct 24;47(2):241–247. doi: 10.1016/0092-8674(86)90446-0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Sassone-Corsi P., Grundström T., Zenke M., Chambon P. Stimulation of in vitro transcription from the SV40 early promoter by the enhancer involves a specific trans-acting factor. EMBO J. 1984 Dec 20;3(13):3129–3133. doi: 10.1002/j.1460-2075.1984.tb02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J. K., Sollner-Webb B. Transcription of Xenopus ribosomal RNA genes by RNA polymerase I in vitro. J Biol Chem. 1982 Dec 10;257(23):14375–14383. [PubMed] [Google Scholar]
- Windle J. J., Sollner-Webb B. Two distant and precisely positioned domains promote transcription of Xenopus laevis rRNA genes: analysis with linker-scanning mutants. Mol Cell Biol. 1986 Dec;6(12):4585–4593. doi: 10.1128/mcb.6.12.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle J., Sollner-Webb B. Upstream domains of the Xenopus laevis rDNA promoter are revealed in microinjected oocytes. Mol Cell Biol. 1986 Apr;6(4):1228–1234. doi: 10.1128/mcb.6.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988 Mar 25;16(5):1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]