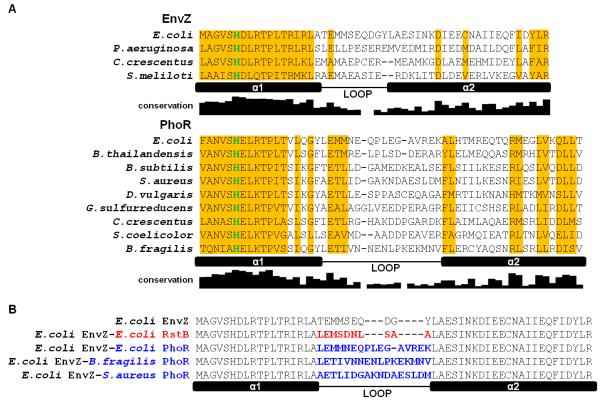
Figure 2. Alignments of EnvZ and PhoR orthologs and loop chimeras.
(A) Sequence alignment of DHp domains from EnvZ and PhoR orthologs characterized in this study. Locations of the two helices in the DHp domain, α1 and α2, and the loop connecting them are indicated. The depicted ortholog loop boundaries are based on sequence alignment to E. coli EnvZ and its loop boundary. The histidine site of phosphorylation is colored in green. Columns where one residue is conserved in more than half of 118 EnvZ or 385 PhoR orthologs are shaded in yellow. The conservation graph below each alignment measures information for each DHp position across all orthologs (see Methods). (B) Alignment of DHp domains of loop chimeras. Sequence changes relative to E. coli EnvZ are highlighted in either red (RstB) or blue (PhoR).