FIGURE 5.
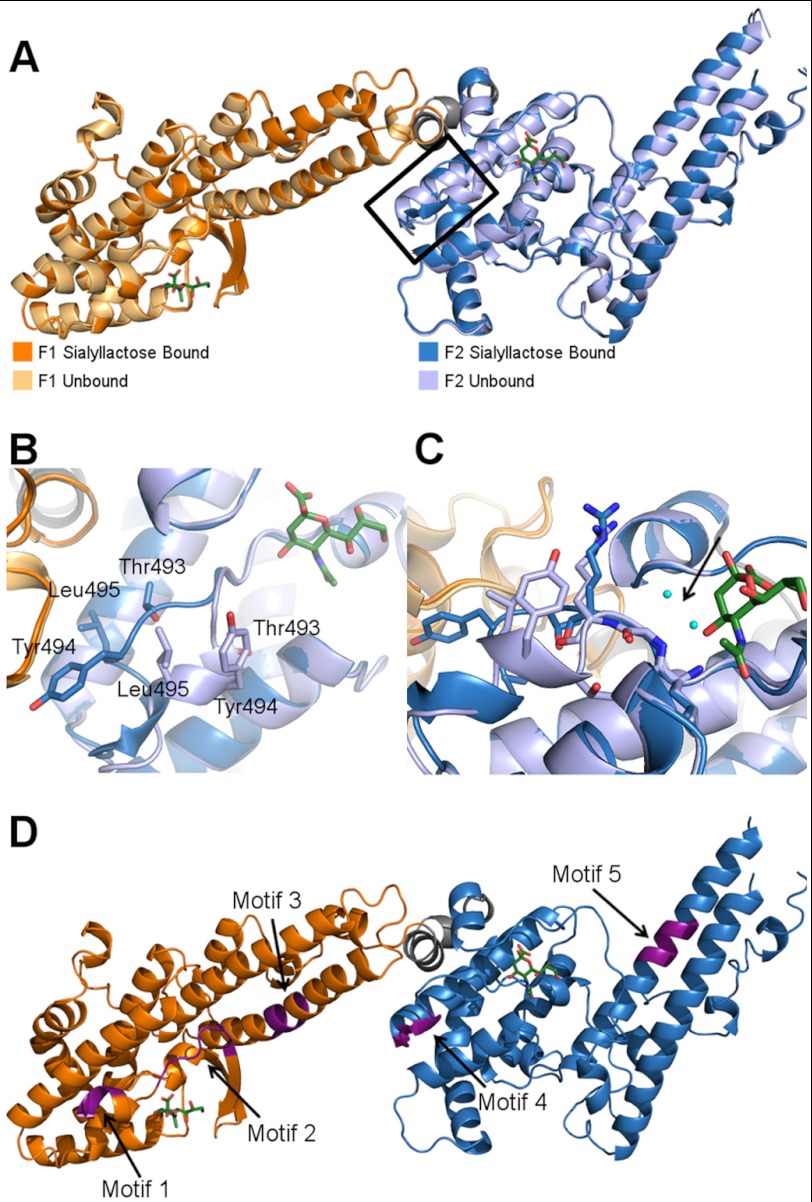
The sialyllactose-bound structure illuminates a glycan-induced structural change and allows for localization of putative sulfate-binding sites. A, overlay of the sialyllactose bound (F1 domain in orange, F2 domain in blue) and unbound (F1 domain in light orange, F2 domain in light blue) structures of RII PfEBA-140. The bound sialic acid molecules are shown in green. The glycan-induced structural change is outlined in black. B, close-up view of residues Thr-493, Tyr-494, and Leu-495, which show a pronounced movement following glycan binding. C, the helix shift in the F2 domain observed upon glycan binding is propagated by sialic acid interference with main chain water molecule interactions. Glycan binding precludes access of two water molecules to the cavity indicated by the black arrow (the waters are present in the apo structure). In the absence of contacts mediated by these water molecules, the helix is destabilized, allowing the movement observed in the bound crystal structure. The water molecules are shown in cyan, and the sialic acid is in green. The bound F2 domain is shown in dark blue, and the unbound F2 domain is in light blue. D, three sulfate-binding motifs in RII PfEBA-140 are adjacent to the sialic acid-binding sites. Two other putative sulfate-binding motifs are distal to the binding sites. The putative sulfate-binding sites are shown in purple and identified with an arrow.