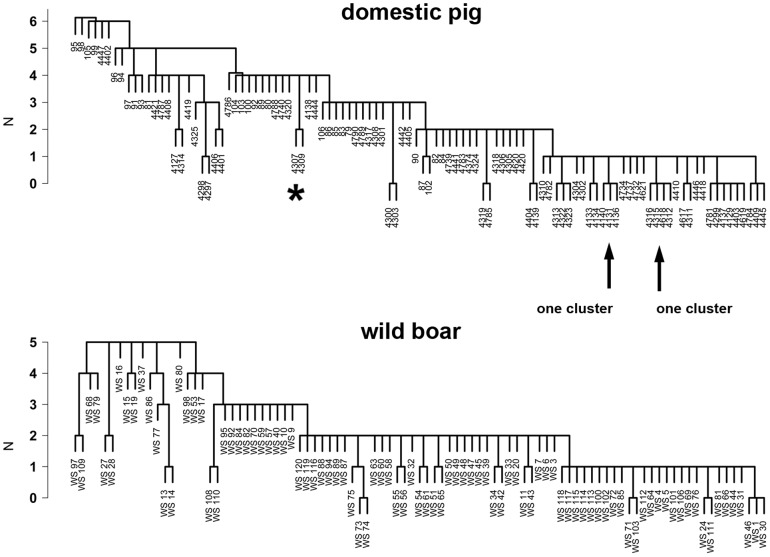
Figure 3. Cluster analysis with virulence-associated genes (VAGs).
Isolates from domestic pigs (n = 104) and from wild boars (n = 93) were clustered according to 9 VAGs typical for E. coli isolated from diarrheic pigs (iVAGs) and 37 VAGs typical for E. coli isolated from extraintestinal disease (eVAGs). Using the manhattan distance measure the distance between strains was computed including a hierarchical cluster analysis using a single linkage method (“friends of friends”) clustering strategy. The minimal difference in the number (N) of genes between clusters of strains to other clusters is described. All isolates of one cluster have identical VAG pattern. Distance of 1 (δN = 1) means that two clusters differed at least in one gene. Clusters which included at least four isolates, which were not different in more than 2 genes, were used to associate virulence gene profiles with adhesion rates or a probiotic effect. There were no associations between a specific cluster and adhesion or a probiotic effect. *: Clustered isolates are identified by strain numbers.