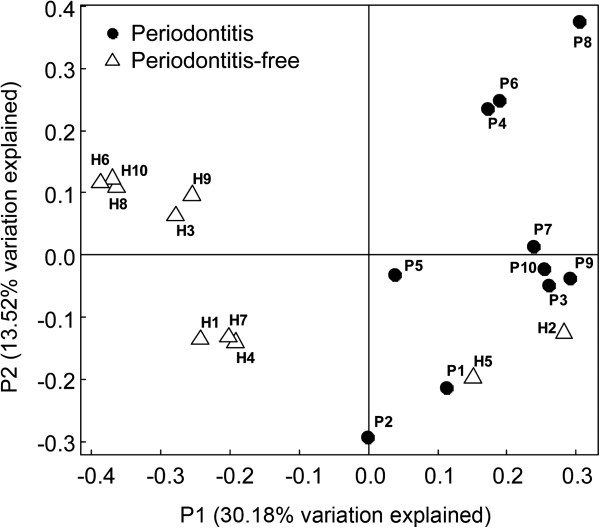
Figure 3.
Principal component analysis (PCoA) of variation amongst the treponeme communities present in each subject. The PCoA was performed using a distance matrix calculated using an unweighted UniFrac algorithm based on a single neighbour-joining (NJ) tree containing all treponeme 16S rRNA gene sequences detected (n = 615). The PCoA scatter plot was constructed using the two principal axes (P1 and P2), which respectively explained 30.18% and 13.52% of the variation between treponeme communities. Black solid circles represent the 10 periodontitis subjects (P1-P10); unshaded triangles represent the 10 periodontitis-free subjects (H1-H10).