Abstract
Until recently, the general perception has been that mutations in protein coding genes are responsible for tumorigenesis. With the discovery of V600EBRAF in about 50% of cutaneous melanomas there was an increased effort to find additional mutations. However, mutations characterized in melanoma to date cannot account for the development of all melanomas. With the discovery of microRNAs as important players in melanomagenesis protein mutations are no longer considered the sole drivers of tumors. Recent research findings have expanded the view for tumor initiation and progression to additional non-coding RNAs. The data suggest that tumorigenesis is likely an interplay between mutated proteins and deregulation of non-coding RNAs in the cell with an additional role of the tumor environment. With the exception of microRNAs, our knowledge of the role of non-coding RNAs in melanoma is in its infancy. Using few examples we will summarize some of the roles of non-coding RNAs in tumorigenesis. Thus, there is a whole world beyond protein coding sequences and microRNAs, which can cause melanoma.
Keywords: melanoma, non-coding RNA, microRNA, epigenetic modifications, alternative splicing
Introduction
Melanoma is one of the few cancers with increasing incidence and deaths over the last decades (Cancer Facts & Figures 2012, Atlanta: American Cancer Society 2012, (1)). There is a massive effort to determine the drivers of cancers and their genetic signatures to understand the biology of tumorigenesis and to identify new therapeutic targets. Although there is no doubt that the tumor microenvironment has a significant influence in tumorigenesis (2–4), we will focus on the role of genetic changes and RNA expression in tumor cells.
Mutations do not explain all tumors
About 50% of cutaneous melanomas harbor a V600EBRAF mutation (5, 6) followed by 15% that have a mutation in NRAS. In familial melanoma, which accounts for about 10% of melanomas, 40% of the patients have a mutation in CDKN2 (7). Additional mutations, like in C-Kit, are found in melanomas in less numerous cases (8). Although the V600EBRAF mutation is the most common mutation in cutaneous melanomas, it is rarely seen in uveal melanomas, those of internal organs (9) or other types of skin cancers. Instead, metastasizing uveal melanomas have a high frequency of BAP1, GNAQ and GNA11 mutations (10, 11).
Despite the high frequency of the above described mutations, they are not sufficient by themselves to induce cancer. For example nevi carry the V600EBRAF mutation without any signs of malignant transformation (12). The same is true for NRAS. A mutation in this gene alone is not sufficient to induce tumors (13). These findings confirm the notion that a second mutation/alteration or even a third is necessary for the transformation of cells, which reflects Knudson’s original two hit theory (14, 15). Using the newest development in 2nd generation sequencing, several additionally mutated genes have been determined in melanomas (reviewed in (8)). The approaches ranged from sequencing just the tyrosine kinome (16), whole exome sequencing (17), whole genome sequencing (18, 19) to transcriptome sequencing (20). Even with these additionally determined mutations we are not able to explain all tumor cases in melanoma. Obviously we are still missing important players in the induction of cellular transformation. One limitation of all the above mentioned studies has been their focus on mutations in protein coding regions of the genome (8). Other mechanisms have been neglected. For example, although the V600EBRAF mutation is common in thyroid cancer, BRAF can also be activated by alternative splicing (21) without the V600E mutation. This alteration would have been most likely missed by solely testing for mutations in the protein coding region. A similar instance can be seen in the resistance to Vemurafenib, a V600EBRAF specific inhibitor (22). Resistance to this drug develops in most treated melanoma patients over time and one of the described resistance mechanism is alternative splicing of V600EBRAF (23). Again alternative splicing can have a similar effect as the introduction of a mutation. Based on these results and others, one has to reconsider if tumorigenesis relies solely on mutations in proteins. This raises the question, if additional genomic and expression modifications occur, which could explain aberrant gene expression in tumor cells?
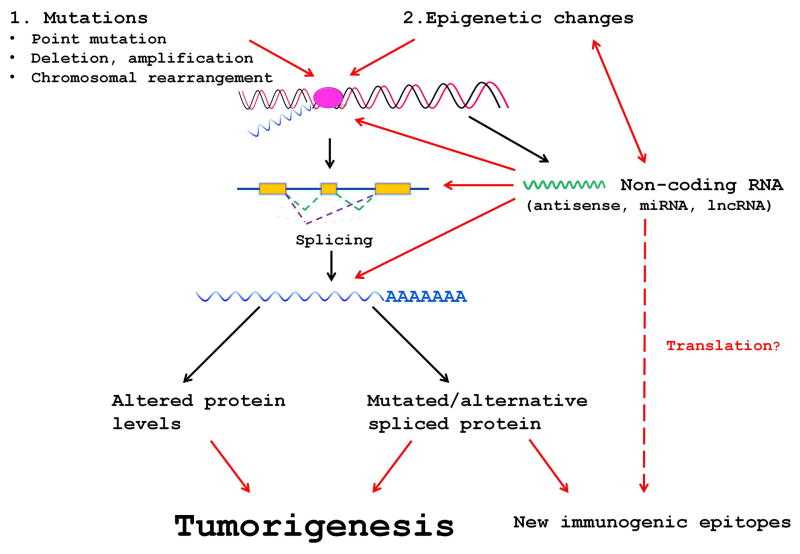
A comprehensive analysis of a malignant melanoma showed that actually the minority of somatic mutations (~1.5% substitution mutations including the UTR region) was found in regions corresponding to mature mRNAs. The majority (~98.5%) of mutations were in other regions of the genome (18). We also know that only a small part of the genome codes for proteins (~2%) and that the majority of RNA is transcribed from non-coding regions (24, 25). The newest data release of ENCODE confirmed that the majority of the genome is transcribed (26). Therefore, our focus on the role of coding genes in tumorigenesis disproportionally favors mRNAs and neglects the influence of non-coding RNAs (ncRNAs). The effects of ncRNAs range from influencing RNA stability, selection of splice variants to general transcription regulation, which either acts in cis by the complementary antisense RNA or in trans like microRNAs (miRNAs). The latter effect includes also long non-coding (lnc) RNAs which can modify the activity of promoters by epigenetic changes (24). An increasing amount of data describe the important role that ncRNAs play in tumors (27). Figure 1 shows different interactions where ncRNAs could influence the expression of mRNAs. Any dysregulation of mRNA by either changing its expression or splice variation could act like an oncogenic event pointing to the significance of ncRNAs and their role in tumorigenesis.
Fig. 1.
Possible interactions of non-coding RNAs with protein expression during tumor induction. Red arrows point to interferences with protein expression by the disturbed expression of non-coding RNAs.
Non-coding RNAs are important in tumorigenesis
microRNAs contribute to melanoma development
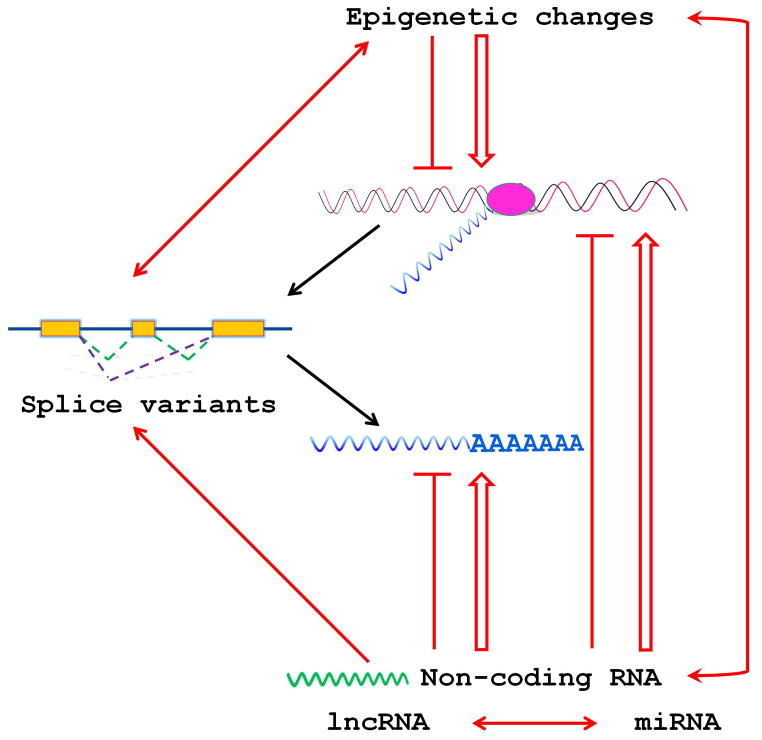
The best studied group of non-coding RNAs are miRNAs, which in most cases decrease targeted mRNA levels (28). For a general review of miRNAs in cancer see reference (29). miRNAs can act as tumor suppressors as well as oncogenes (30) and there is no doubt about their role in tumorigenesis (31). In tumors the biogenesis of miRNAs is disturbed (32, 33), which will alter the expression levels of miRNAs and ultimately the expression of genes regulated by miRNAs (32). In melanoma, the miRNAome (34) has been determined and the role of miRNAs in melanomas has been reviewed (35–37). miRNAs are involved in all steps of tumorigenesis from initiation (38) to metastasis (39, 40). Melanoma subtypes differ in their miRNA signatures (41), which can serve as a prognostic biomarker (42). Additionally, miRNAs not only regulate mRNAs but also other ncRNAs (43, 44) and they themselves are epigenetically regulated (45). This places ncRNAs in a wider, multilayer regulatory network of transcriptional and translational control (Fig. 2).
Fig. 2.
Simplified map of interactions in the non-coding RNA network itself and with mRNA expression. The symbols mean
 inhibition,
inhibition,
 stimulation,
stimulation,
 mutual interaction.
mutual interaction.
Do lncRNAs have a role in melanomagenesis?
Besides miRNAs, there are also lncRNAs (46). As implied by their name, they are larger than miRNAs with a minimum length of 200bp and up to several kilobases. One of the best studied members is XIST, which is involved in the inactivation of the X chromosome (47). lncRNAs are divided in three major groups: long intergenic non-coding RNAs (lincRNA), which are located away from protein coding regions, antisense RNAs (asRNA), which are transcribed in reverse orientation and overlap with a known gene, and intronic lncRNAs. The groups have partly overlapping activities. For example, members of each group regulate epigenetic modifications, however only the transcription of asRNAs directly interferes with transcription of its corresponding sense counterpart by polymerase stalling. The different functions attributed to lncRNAs (reviewed in (48, 49)) include control of pluripotency and differentiation in stem cells (50), setting of epigenetic marks (51), and functioning as enhancer RNAs (52). One subgroup of intron-derived lncRNAs, the sno-lncRNAs, is associated with changes in splicing (53). Although lncRNAs are expressed in normal cells with tissue-specific expression patterns, aberrant expression occurs in tumors (54) and they play roles in cancer progression (55). Based on the expanding knowledge of their regulatory influence lncRNAs may represent “a new frontier of translational research” in cancer (56). There is very limited information on the role of specific lncRNAs in melanoma. In 2011, Khaitan et al.(57) showed that the lncRNA SPRY4-IT1 modulates cell growth and differentiation and its knock-down increased apoptosis. Flockhart et al.(58) described the effect of V600EBRAF on the expression of about 100 lncRNAs.
The number of known lncRNAs is still increasing. The subclass lincRNAs alone contains over 8,000 members (59) with different activities. One of the described functions of lnc RNAs is setting of epigenetic marks. For example, the lncRNA HOTAIR is involved in the setting of epigenetic marks by the polycomb repressive complex 2 and its expression levels are increased in breast tumors. A high expression level implies a poor prognosis for metastasis and survival in breast tumor patients (60). It is well established that epigenetic changes occur in tumors (61, 62), and that these changes play a role in melanomas as well (63). Interestingly, conditions which are indirectly linked to the onset of tumors, such as stress (64, 65) and age (66), induce epigenetic changes. Issa and Garber suggested the presence of an epigenetic predisposition to cancer (67). Therefore, epigenetic changes induced by dysregulation of ncRNAs may act like an oncogenic event. One has to emphasize that different non-coding RNAs, lincRNAs, asRNAs (68) as well as miRNAs can induce epigenetic changes, but at the same time miRNAs are regulated by epigenetic changes (45).
The next group of lncRNAs are asRNAs, which not only contain non-coding RNAs but also coding RNAs although to a lesser extent (e.g. FGF-2 and its asRNA FGF-AS (69)). AsRNAs are relatively common (70, 71). They regulate the expression of their corresponding sense genes by different mechanisms (72), like influencing sense RNA stability, epigenetic changes and alternative splicing (73, 74). The idea that splicing is a factor in tumorigenesis is not new (75, 76), but the study and detection of splice variants was previously hampered by technical limitations. With increasing numbers of high-throughput RNA sequencing studies completed (reviewed in (77)), we will get a more complete list of splice variations (78) between normal and diseased tissues and thus a better understanding of their role in cancer, including melanoma. In mice, melanocytes and melanomas differ in splice variants (79). Even more astonishing is the observation that breast cancer cells grown in 2D or 3D culture differ in their splice variants (80), which suggests that alternative splicing can occur in response to subtle changes in the microenvironment. In melanoma, different splice isoforms of MITF, the master regulator of melanocyte development, have been described (81) as well as aberrant splicing of the tumor suppressor Bin1 (82). Splicing even plays a role in resistance to cancer treatments such as Vemurafenib (23). Based on these findings, targeting specific splice variants by antisense oligonucleotides has been suggested for cancer therapy (83) and splice variants have been proposed as cancer biomarkers (84).
Alternative splicing is not an isolated incident. Splice variants of DNA methyltransferases (85) differ in their activity, which may result in epigenetic changes. At the same time epigenetic changes regulate alternative splicing (86). Additionally, changes in the 3’UTR of mRNAs due to alternative splicing can alter the recognition by miRNAs (87). Thus, alternative splicing is part of a larger interacting network and is influenced by ncRNAs (88).
Minor modulation of mRNA levels can have a significant effect
In any discussion about the role of ncRNAs, one has to be aware that already small expression level changes can have an effect. One of the best examples is the tumor suppressor PTEN (89). Pandolfi’s group (90, 91) has demonstrated that small changes in the expression levels of PTEN are sufficient to achieve an effect in tumor progression, thus, complete deletion is not necessary. It is known that asRNA levels influence sense RNA levels. Epigenetic changes are another way to vary RNA expression. The discovery of ceRNAs (competing endogenous mRNAs), which influence mRNA levels by competing for miRNAs, adds one more mechanism to modulate the expression level of a certain gene (92–94).
Are non-coding RNAs the new frontier in melanoma research?
Biological and bioinformatics analyses of non-coding regions are still in their infancy, with the exception of the well-established miRNA field. The cancer genome atlas is just the beginning and much research remains to be done. We will gather much information on exon sequences, genomic deletions and amplifications, but like first-graders we still lack the knowledge to fully comprehend the information contained in the whole genomic sequence. We still do not know how to interpret the sequences and possible mutations of non-coding RNAs. In contrast to protein coding sequences, no reference sequences exist for those regions to define wild type versus mutated versions. Because the functions of most ncRNAs are unknown, no assays are available to determine, which sequence changes influence their function.
Analyzing each single compartment (protein, mRNA, ncRNA) and their interactions is already a major challenge. However, the analyses of the key players are further hindered by the fact that splice variants, epigenetic changes and ncRNAs interact and influence each other. For example, histone modifications regulate alternative splicing (86) and conversely, splice variants of DNA methyltransferases result in epigenetic changes (85). All these data stress the fact that control of mRNA expression (transcription factors, translation, etc.) is overlaid and controlled by an interacting network of ncRNAs (Fig. 2). Any disturbance at any level, including ncRNA interactions, will eventually have consequences at the protein level. This will either be reflected in the quantity of protein or in introduced modifications (e.g. alternative splicing).
Summary and perspectives
The discovery of V600EBRAF was a blessing and a curse for melanoma research. It gave the melanoma community a new target with impressive therapeutic results for patients, but at the same time resulted in neglect to study other areas, such as ncRNAs. We are aware that splice variants and asRNAs were in vogue some time ago and we are not the first to suggest studying them more carefully. It is time to return to old ideas using new tools (e.g. RNAseq). The overall fixation on mutations in coding genes as a cause of transformation limits us. It is time to combine both worlds, the coding as well as the non-coding RNAs, in our studies to understand the biology of cancer and to determine new targets in our battle with cancer. NcRNAs will be valid targets for tumor treatment and with the development of new RNA therapeutics we may be able to target ncRNAs more efficiently than with conventional antisense oligonucleotides (95). Additionally, ncRNAs may not be only direct targets. The results of Ingolia et al.(96) suggest that translation may occur from ncRNAs, which could provide new epitopes to develop tumor specific immunotherapies. In our own research, we have identified tumor specific T cell antigens encoded by asRNAs. Table 1 shows some examples of conceivable applications based on results gained by studying ncRNAs.
Table 1.
Conceivable applications based on non-coding RNAs.
|
|
We are well aware that this viewpoint omits important aspects of tumor drivers such as the influence of the tumor microenvironment (2–4), and other modifications such as alternative initiation of translation on non-AUG codons (97) or RNA editing (98). Considering all the possible modifications which can occur in tumor cells, one may feel overwhelmed whether we will ever come close to full understanding and conquering cancer. On the other hand, with all these multilayer regulatory networks we may find new “weak spots” of tumors for intervention. We provided only a glimpse into the world of non-coding RNAs and their possible roles in tumorigenesis. We hope to inspire readers to expand their view beyond mutated proteins and to include non-coding RNAs in their studies. We apologize to all colleagues whose work we could not cite due to space restrictions.
Acknowledgments
The research in the laboratory of M.H. is funded by NIH grants CA076674, CA025874 and CA047159.
Footnotes
Conflict of interests
The authors have declared no conflict of interests.
Both authors contributed to the manuscript and approved the final version.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva J, Herlyn M. Melanoma and the tumor microenvironment. Curr Oncol Rep. 2008;10:439–446. doi: 10.1007/s11912-008-0067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164:776–784. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66:9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 8.Walia V, Mu EW, Lin JC, Samuels Y. Delving into somatic variation in sporadic melanoma. Pigment Cell Melanoma Res. 2012;25:155–170. doi: 10.1111/j.1755-148X.2012.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong CW, Fan YS, Chan TL, et al. BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J Clin Pathol. 2005;58:640–644. doi: 10.1136/jcp.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R, Angelini S, Snellman E, Hemminki K. BRAF mutations are common somatic events in melanocytic nevi. J Invest Dermatol. 2004;122:342–348. doi: 10.1046/j.0022-202X.2004.22225.x. [DOI] [PubMed] [Google Scholar]
- 13.Lin K, Baritaki S, Militello L, Malaponte G, Bevelacqua Y, Bonavida B. The Role of B-RAF Mutations in Melanoma and the Induction of EMT via Dysregulation of the NF-kappaB/Snail/RKIP/PTEN Circuit. Genes Cancer. 2010;1:409–420. doi: 10.1177/1947601910373795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knudson AG. Hereditary cancer: Two hits revisited. Journal of Cancer Research and Clinical Oncology. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson IP, Roylance R, Houlston RS. Two hits revisited again. J Med Genet. 2001;38:81–85. doi: 10.1136/jmg.38.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baitei EY, Zou M, Al-Mohanna F, et al. Aberrant BRAF splicing as an alternative mechanism for oncogenic B-Raf activation in thyroid carcinoma. J Pathol. 2009;217:707–715. doi: 10.1002/path.2496. [DOI] [PubMed] [Google Scholar]
- 22.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 25.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 26.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SK, Calin GA. Non-coding RNAs and cancer: new paradigms in oncology. Discov Med. 2011;11:245–254. [PubMed] [Google Scholar]
- 28.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 31.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis-Dusenbery BN, Hata A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes Cancer. 2010;1:1100–1114. doi: 10.1177/1947601910396213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Stark MS, Tyagi S, Nancarrow DJ, et al. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010;5:e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glud M, Gniadecki R. MicroRNAs in the pathogenesis of malignant melanoma. J Eur Acad Dermatol Venereol. 2012 doi: 10.1111/j.1468-3083.2012.04579.x. [DOI] [PubMed] [Google Scholar]
- 36.Philippidou D, Schmitt M, Moser D, et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 37.Mueller DW, Bosserhoff AK. The evolving concept of ‘melano-miRs’-microRNAs in melanomagenesis. Pigment Cell Melanoma Res. 2010;23:620–626. doi: 10.1111/j.1755-148X.2010.00734.x. [DOI] [PubMed] [Google Scholar]
- 38.Streicher KL, Zhu W, Lehmann KP, et al. A novel oncogenic role for the miRNA-506–514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558–1570. doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 39.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Mazar J, DeYoung K, Khaitan D, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One. 2010;5:e13779. doi: 10.1371/journal.pone.0013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan E, Patel R, Nallur S, et al. MicroRNA signatures differentiate melanoma subtypes. Cell Cycle. 2011;10:1845–1852. doi: 10.4161/cc.10.11.15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segura MF, Belitskaya-Levy I, Rose AE, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi JJ. A novel nuclear miRNA mediated modulation of a non-coding antisense RNA and its cognate sense coding mRNA. EMBO J. 2011;30:4340–4341. doi: 10.1038/emboj.2011.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011;585:2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang XQ, Crutchley JL, Dostie J. Shaping the Genome with Non-Coding RNAs. Curr Genomics. 2011;12:307–321. doi: 10.2174/138920211796429772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 52.Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin QF, Yang L, Zhang Y, et al. Long Noncoding RNAs with snoRNA Ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 54.Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 58.Flockhart RJ, Webster DE, Qu K, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schinke C, Mo Y, Yu Y, et al. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–265. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sigalotti L, Covre A, Fratta E, et al. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnstone SE, Baylin SB. Stress and the epigenetic landscape: a link to the pathobiology of human diseases? Nat Rev Genet. 2010;11:806–812. doi: 10.1038/nrg2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maegawa S, Hinkal G, Kim HS, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Issa JP, Garber JE. Time to think outside the (genetic) box. Cancer Prev Res (Phila) 2011;4:6–8. doi: 10.1158/1940-6207.CAPR-10-0348. [DOI] [PubMed] [Google Scholar]
- 68.Su WY, Xiong H, Fang JY. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem Biophys Res Commun. 2010;396:177–181. doi: 10.1016/j.bbrc.2010.04.147. [DOI] [PubMed] [Google Scholar]
- 69.Knee RS, Pitcher SE, Murphy PR. Basic fibroblast growth factor sense (FGF) and antisense (gfg) RNA transcripts are expressed in unfertilized human oocytes and in differentiated adult tissues. Biochem Biophys Res Commun. 1994;205:577–583. doi: 10.1006/bbrc.1994.2704. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, Sun M, Kent WJ, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 72.Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 73.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrissy AS, Griffith M, Marra MA. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011;21:1203–1212. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu A, Fu XD. Splicing oncogenes. Nat Struct Mol Biol. 2007;14:174–175. doi: 10.1038/nsmb0307-174. [DOI] [PubMed] [Google Scholar]
- 76.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 79.Watahiki A, Waki K, Hayatsu N, et al. Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nat Methods. 2004;1:233–239. doi: 10.1038/nmeth719. [DOI] [PubMed] [Google Scholar]
- 80.Li C, Kato M, Shiue L, Shively JE, Ares M, Jr, Lin RJ. Cell type and culture condition-dependent alternative splicing in human breast cancer cells revealed by splicing-sensitive microarrays. Cancer Res. 2006;66:1990–1999. doi: 10.1158/0008-5472.CAN-05-2593. [DOI] [PubMed] [Google Scholar]
- 81.Primot A, Mogha A, Corre S, et al. ERK-regulated differential expression of the Mitf 6a/b splicing isoforms in melanoma. Pigment Cell Melanoma Res. 2010;23:93–102. doi: 10.1111/j.1755-148X.2009.00652.x. [DOI] [PubMed] [Google Scholar]
- 82.Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast GC. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci U S A. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bartel F, Harris LC, Wurl P, Taubert H. MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res. 2004;2:29–35. [PubMed] [Google Scholar]
- 84.Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 85.Van Emburgh BO, Robertson KD. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res. 2011;39:4984–5002. doi: 10.1093/nar/gkr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durand C, Roeth R, Dweep H, et al. Alternative splicing and nonsense-mediated RNA decay contribute to the regulation of SHOX expression. PLoS One. 2011;6:e18115. doi: 10.1371/journal.pone.0018115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 90.Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carracedo A, Alimonti A, Pandolfi PP. PTEN level in tumor suppression: how much is too little? Cancer Res. 2011;71:629–633. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sumazin P, Yang X, Chiu HS, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Touriol C, Bornes S, Bonnal S, et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 98.Gallo A, Galardi S. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 2008;5:135–139. doi: 10.4161/rna.5.3.6739. [DOI] [PubMed] [Google Scholar]