Abstract
Background
The prevalence and type of plasmids, resistance genes and integrons carried by two collections of multiresistant E. coli producing or not extended-spectrum β-lactamases have been compared. Rep-PCR was used to determine the clonal relationship of the organisms. Plasmids were classified according to their incompatibility. Class 1 and Class 2 integrons and antibiotic resistance genes were analysed by PCR and sequencing.
Results
Both collections of organisms contained a large diversity of unrelated strains with some clones distributed in both groups of isolates. Large plasmids were identified in the two groups of organisms. Plasmids with replicons repK and repColE were more frequent among ESBL-producing isolates, while repFIA, repFII and repA/C replicons were more frequent in isolates lacking ESBL. Conjugative plasmids with repK and repA/C replicons coded for CTX-M-14 and CMY-2 β-lactamases, respectively. No significant differences were observed in the distribution of class 1 and class 2 integrons among multiresistant E. coli producing or not ESBL, and dfrA17-ant(3″)-Ie was the cassette arrangement most commonly found.
Conclusions
In the concrete temporal and geographical context of this study, multiresistant E. coli producing ESBL or other mechanisms of resistance were largely clonally diverse and present some differences in the types of harboured plasmids. Still, some clones were found in both ESBL-producing and –lacking isolates.
Keywords: E. coli, Plasmids, ESBL, Multiresistance
Background
A rapid dissemination of Escherichia coli and others enterobacteria producing extended-spectrum beta-lactamases (ESBLs) has been reported in many European countries, including Spain, and is a matter of major concern [1,2]. The blaCTX-M genes are becoming the most prevalent ESBLs encountered in Enterobacteriaceae[2]. The prevalent blaCTX-M-type genes in Europe have been identified as blaCTX-M-1, blaCTX-M-3, blaCTX-M-9, blaCTX-M-14 and blaCTX-M-15[2]. Infections caused by enterobacteria producing ESBL are associated with increased morbidity, mortality, and health-care associated costs [3].
During recent years, extensive characterization of plasmid families (usually by replicon typing [4,5] or, more recently, by relaxase identification [6]) has provided additional information on epidemiological aspects of transmissible antimicrobial resistance [7]. Many of these studies have focused on E. coli producing ESBL [8,9]. From these studies, some plasmid families were demonstrated to be largely prevalent in Enterobacteriaceae, emerging in association with specific ESBL genes. For instance, the blaCTX-M-15 and blaCTX-M-3 genes have often been located on plasmids belonging to the IncF group [10] and IncL/M family [11] respectively, in different countries [12-16]. It would be interesting to know if in a specific geographical area, plasmid-mediated antimicrobial resistance in multiresistant E. coli producing ESBL is due to plasmids of the same incompatibility group(s) as those present in other multirresistant isolates not producing ESBL.
The objective of this study was to determine whether the clonal variability and content of plasmids, resistance genes and integrons of clinical isolates of E. coli producing ESBL (Ec-ESBL) were similar or not to those of E. coli isolates lacking ESBL (Ec-MRnoB) isolated in the same geographical area and period.
Results
Phenotype of resistance and clonal relationship
MIC50 and MIC90 values of the different agents against the two groups of multiresistant E. coli are presented in Table 1. All Ec-ESBL were susceptible to cefotetan, imipenem, meropenem, amikacin and tigecycline. According to the EUCAST [17], all Ec-ESBL isolates were resistant to cefotaxime, 96% to cefepime, 96% to aztreonam and 23% to ceftazidime (Table 1). Moreover, 39% of the isolates belonging to the Ec-ESBL collection were co-resistant to quinolones, tetracycline and trimethoprim-sulfamethoxazole.
Table 1.
MIC range, MIC50 (mg/L) and MIC90 (mg/L) values of 21 antimicrobial agents for Ec-ESBL and Ec-MRnoB isolates, and percentages of resistant isolates to the indicated agents (EUCAST breakpoints, except where indicated)
| |
Ec-ESBL (N=100) |
Ec-MRnoB (N=100) |
||||||
|---|---|---|---|---|---|---|---|---|
| Agent | Range | MIC50 | MIC90 | %R | Range | MIC50 | MIC90 | %R |
|
Amoxicillin |
64-≥256 |
≥256 |
≥256 |
100 |
2-≥256 |
≥256 |
≥256 |
99 |
|
Amoxicillin-Clavulanic acid |
8-16 |
4 |
8 |
41 |
8-≥128 |
8 |
32 |
83 |
|
Piperacillin |
32-≥64 |
≥64 |
≥64 |
100 |
≤16-≥64 |
≥64 |
≥64 |
84 |
|
Piperacillin-Tazobactam |
≤0.5-≥256 |
4 |
16 |
16 |
≤0.25-≥256 |
2 |
4 |
6 |
|
Cefoxitina |
≤2-32 |
≤2 |
8 |
5 |
≤2-≥32 |
4 |
8 |
8 |
|
Cefotetana |
≤1-8 |
≤1 |
≤1 |
0 |
≤1-≥32 |
≤1 |
≤1 |
2 |
|
Cefotaxime |
32-≥128 |
≥128 |
≥128 |
100 |
≤0.5-128 |
≤0.5 |
≤0.5 |
5 |
|
Ceftazidime |
≤0.5-≥128 |
2 |
16 |
23 |
≤0.5-128 |
≤0.5 |
≤0.5 |
8 |
|
Cefepime |
≤1-≥32 |
≥32 |
≥32 |
96 |
≤1-4 |
≤1 |
<1 |
1 |
|
Aztreonam |
2-≥64 |
16 |
≥64 |
96 |
≤0.5-32 |
≤0.5 |
≤0.5 |
6 |
|
Imipenem |
≤0.5-1 |
≤0.5 |
≤0.5 |
0 |
≤0.5-2 |
≤0.5 |
≤0.5 |
0 |
|
Meropenem |
≤0.5 |
≤0.5 |
≤0.5 |
0 |
≤0.5 |
≤0.5 |
≤0.5 |
0 |
|
Gentamicin |
≤0.5-≥256 |
1 |
32 |
19 |
0.5-≥256 |
64 |
256 |
96 |
|
Tobramycin |
0.5-256 |
1 |
8 |
17 |
≤0.25-256 |
8 |
32 |
89 |
|
Amikacin |
≤0.5-8 |
2 |
4 |
0 |
1-8 |
2 |
4 |
0 |
|
Nalidixic acida |
1-≥256 |
≥256 |
≥256 |
89 |
1-≥256 |
≥256 |
≥256 |
98 |
|
Ciprofloxacina |
≤0.5-≥256 |
16 |
128 |
74 |
≤0.25-≥256 |
32 |
128 |
93 |
|
Tetracyclinea |
0.5-≥256 |
256 |
256 |
80 |
≤0.25-256 |
256 |
256 |
84 |
|
Doxycyclinea |
≤0.5-128 |
16 |
64 |
76 |
≤0.25-128 |
32 |
64 |
79 |
|
Tigecyclineb |
≤0.5-1 |
≤0.5 |
≤0.5 |
0 |
≤0.25-0.5 |
≤0.25 |
≤0.25 |
0 |
| Trimethoprim-Sulfamethoxazole | ≤0.5-≥32 | ≥32/608 | ≥32/608 | 67 | ≤0.5-≥32 | ≥32/608 | ≥32/608 | 98 |
a CLSI 2011 breakpoints. b FDA breakpoints.
All Ec-MRnoB were susceptible to imipenem, meropenem, amikacin and tigecycline. Eight isolates in this collection were resistant to at least one extended-spectrum cephalosporin (ceftazidime) (Table 1). The most frequent phenotype of resistance observed among the selected Ec-MRnoB isolates included resistance to β-lactams (amoxicillin), aminoglycosides [gentamicin alone or (more often) associated to tobramycin], quinolones (nalidixic acid alone or associated to ciprofloxacin), tetracyclines (tetracycline alone or associated to doxycycline) and trimethoprim-sulfamethoxazole, occurring in 50% of the studied isolates. All other possible combinations of co-resistances among the selected isolates represented no more than 5% of the isolates.
Most Ec-ESBL were of phylogroup B1 (38%), followed by groups A (32%), D (22%) and B2 (8%). In contrast, the most frequent phylogenetic group of Ec-MRnoB was D (46%), followed by groups A (25%), B2 (17%) and B1 (12%).
The 100 Ec-ESBL isolates were grouped in 66 Rep-PCR patterns. In this group, only 2 Rep-PCR patterns included 5 or more isolates: patterns XXXI (n=6, phylogenetic group A) and XXII (n=5; B1). The remaining patterns contained 2 to 4 isolates (16 Rep-PCR patterns) or single isolates (48 Rep-PCR patterns). Lower clonal variability was noted among the Ec-MRnoB, which were grouped into 40 Rep-PCR patterns. Three patterns included 5 or more isolates: I-NB (n=18, phylogenetic group D), II-NB (n=14; B2) and XXIII-NB (n=8; D). Fifteen patterns included 2 to 4 isolates, and the remaining 22 patterns corresponded to single isolates.
Comparison of Rep-PCR patterns corresponding to isolates of the two E. coli collections showed the presence of Ec-ESBL (5 Rep-PCR patterns corresponding to 11 isolates) and Ec-MRnoB (4 Rep-PCR patterns corresponding to 30 isolates) with the same pattern. PFGE analysis confirmed this relationship, with isolates presenting identical or similar [less than 5 bands of difference] PFGE patterns. In addition, clonally related isolates (determined by Rep-PCR/PFGE) were included in the same ST defined by MLST (Figure 1).
Figure 1.
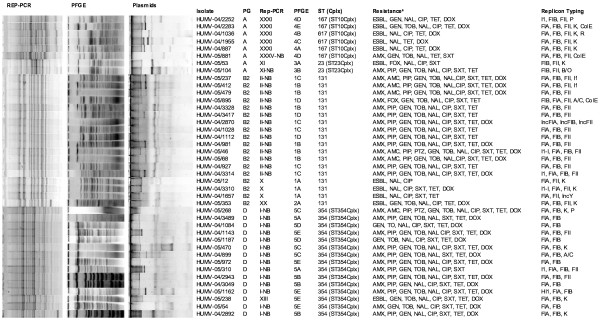
Clonal relationship and plasmid profile of related isolates from both collections. aESBL= extended-spectrum β-lactamases; AMX=amoxicillin, PIP= piperacillin; AMC=amoxicillin-clavulanic acid; CAZ=ceftazidime, FOX=cefoxitin; GEN=gentamicin, TOB=tobramycin, NAL=nalidixic acid, CIP=ciprofloxacin, TET= tetracycline, SXT= trimethoprim.-sulphamethoxazole.
Twenty-four ST were identified among the isolates selected for the conjugation assay (Table 2), of which seven types included both Ec-ESBL and Ec-MRnoB, while 11 and 6 types included either Ec-ESBL or Ec-MRnoB, respectively. Three new ST (ST2292, ST2294 and ST2295) were first identified among our Ec-ESBL isolates.
Table 2.
Clonal complexes (Cplx) and sequence types (ST) of 40 multiresistant E. coli producing ESBL (Ec-ESBL, n=20) or not (EcMRnoB, n=20) selected for the conjugation assay
| Isolate | E. coli collection | ST | Cplx | adk | fumC | gyrB | icd | mdh | purA | recA | Rep-PCR | Nª |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HUMV-05/374 |
Ec-MRnoB |
ST1011 |
|
6 |
4 |
159 |
44 |
112 |
1 |
17 |
III-NB |
3 |
| HUMV-04/2539 |
Ec-ESBL |
ST117 |
|
20 |
45 |
41 |
43 |
5 |
32 |
2 |
V |
2 |
| HUMV-05/21 |
Ec-MRnoB |
ST117 |
|
20 |
45 |
41 |
43 |
5 |
32 |
2 |
XXIII-NB |
8 |
| HUMV-04/1119 |
Ec-MRnoB |
ST1210 |
|
187 |
11 |
4 |
8 |
8 |
8 |
2 |
IV-NB |
3 |
| HUMV-04/925 |
Ec-MRnoB |
ST1210 |
|
187 |
11 |
4 |
8 |
8 |
8 |
2 |
VI-NB |
4 |
| HUMV-04/3310 |
Ec-ESBL |
ST131 |
|
53 |
40 |
47 |
13 |
36 |
28 |
29 |
X |
3 |
| HUMV-05/46 |
Ec-MRnoB |
ST131 |
|
53 |
40 |
47 |
13 |
36 |
28 |
29 |
II-NB |
14 |
| HUMV-05/895 |
Ec-MRnoB |
ST131 |
|
53 |
40 |
47 |
13 |
36 |
28 |
29 |
II-NB |
14 |
| HUMV-04/2296 |
Ec-MRnoB |
ST131 |
|
53 |
40 |
47 |
13 |
36 |
28 |
29 |
XII-NB |
1 |
| HUMV-04/2830 |
Ec-ESBL |
ST155 |
ST155Cplx |
6 |
4 |
14 |
16 |
24 |
8 |
|
XXII |
5 |
| HUMV-04/2103 |
Ec-ESBL |
ST156 |
ST156Cplx |
6 |
29 |
32 |
16 |
11 |
8 |
44 |
XXXVII |
3 |
| HUMV-04/2283 |
Ec-ESBL |
ST167 |
ST10Cplx |
10 |
11 |
4 |
8 |
8 |
13 |
2 |
XXXI |
6 |
| HUMV-05/868 |
Ec-MRnoB |
ST167 |
ST10Cplx |
10 |
11 |
4 |
8 |
8 |
13 |
2 |
VI-NB |
4 |
| HUMV-05/881 |
Ec-MRnoB |
ST167 |
ST10Cplx |
10 |
11 |
4 |
8 |
8 |
13 |
2 |
XXXV-NB |
1 |
| HUMV-04/2487 |
Ec-ESBL |
ST224 |
|
6 |
4 |
33 |
16 |
11 |
8 |
6 |
XXIV |
2 |
| HUMV-04/3181 |
Ec-MRnoB |
ST224 |
|
6 |
4 |
33 |
16 |
11 |
8 |
6 |
X-NB |
3 |
| HUMV-04/3218 |
Ec-MRnoB |
ST224 |
|
6 |
4 |
33 |
16 |
11 |
8 |
6 |
XVII-NB |
1 |
| HUMV-04/1412 |
Ec-ESBL |
ST2292 |
|
6 |
65 |
15 |
18 |
9 |
8 |
6 |
LIII |
1 |
| HUMV-04/1391 |
Ec-ESBL |
ST2296 |
|
8 |
7 |
1 |
8 |
6 |
8 |
7 |
XXII |
5 |
| HUMV-05/284 |
Ec-ESBL |
ST2295 |
|
8 |
7 |
14 |
8 |
8 |
8 |
6 |
XVII |
3 |
| HUMV-04/3168 |
Ec-MRnoB |
ST23 |
ST23Cplx |
6 |
4 |
12 |
1 |
20 |
13 |
7 |
XXXIX-NB |
2 |
| HUMV-05/268 |
Ec-MRnoB |
ST354 |
ST354Cplx |
85 |
88 |
78 |
29 |
59 |
58 |
62 |
I-NB |
18 |
| HUMV-05/310 |
Ec-MRnoB |
ST354 |
ST354Cplx |
85 |
88 |
78 |
29 |
59 |
58 |
62 |
I-NB |
18 |
| HUMV-05/281 |
Ec-ESBL |
ST359 |
|
43 |
41 |
15 |
90 |
11 |
8 |
6 |
XIV |
4 |
| HUMV-05/508 |
Ec-ESBL |
ST359 |
|
43 |
41 |
15 |
90 |
11 |
8 |
6 |
XIV |
4 |
| HUMV-04/1087 |
Ec-ESBL |
ST362 |
|
62 |
100 |
17 |
31 |
5 |
5 |
4 |
XXIII |
2 |
| HUMV-04/3096 |
Ec-ESBL |
ST393 |
ST31Cplx |
18 |
106 |
17 |
6 |
5 |
5 |
4 |
III |
4 |
| HUMV-05/631 |
Ec-MRnoB |
ST393 |
ST31Cplx |
18 |
106 |
17 |
6 |
5 |
5 |
4 |
XV-NB |
3 |
| HUMV-04/2833 |
Ec-ESBL |
ST538 |
ST538Cplx |
13 |
40 |
19 |
13 |
36 |
28 |
30 |
IV |
3 |
| HUMV-04/1630 |
Ec-ESBL |
ST57 |
ST350Cplx |
6 |
31 |
5 |
28 |
1 |
1 |
2 |
XXXIX |
2 |
| HUMV-05/190 |
Ec-ESBL |
ST617 |
ST10Cplx |
10 |
11 |
4 |
8 |
8 |
13 |
73 |
II |
3 |
| HUMV-04/3317 |
Ec-ESBL |
ST617 |
ST10Cplx |
10 |
11 |
4 |
8 |
8 |
13 |
73 |
XXXI |
6 |
| HUMV-04/2942 |
Ec-ESBL |
ST641 |
ST86Cplx |
9 |
6 |
33 |
131 |
24 |
8 |
7 |
XVI |
2 |
| HUMV-04/1012 |
Ec-ESBL |
ST648 |
|
92 |
4 |
87 |
96 |
70 |
58 |
2 |
LVIII |
2 |
| HUMV-05/630 |
Ec-MRnoB |
ST648 |
|
92 |
4 |
87 |
96 |
70 |
58 |
2 |
XXXI-NB |
1 |
| HUMV-04/1002 |
Ec-MRnoB |
ST69 |
ST69Cplx |
21 |
35 |
27 |
6 |
5 |
5 |
4 |
IX-NB |
4 |
| HUMV-04/1725 |
Ec-ESBL |
ST88 |
ST23Cplx |
6 |
4 |
12 |
1 |
20 |
12 |
7 |
XXXVI |
2 |
| HUMV-05/569 |
Ec-MRnoB |
ST88 |
ST23Cplx |
6 |
4 |
12 |
1 |
20 |
12 |
7 |
XXIV-NB |
4 |
| HUMV-05/523 |
Ec-MRnoB |
ST88 |
ST23Cplx |
6 |
4 |
12 |
1 |
20 |
12 |
7 |
XXX-NB |
1 |
| HUMV-04/979 | Ec-MRnoB | ST93 | ST168Cplx | 6 | 11 | 4 | 10 | 7 | 8 | 6 | XXXVII-NB | 1 |
aTotal number of isolates belonging to the indicated Rep-PCR pattern.
Plasmid profile
One or more plasmids were observed in organisms of the two E. coli collections. For the 69 Ec-ESBL isolates the most frequently detected replicons were repFIB (64/69, 92.8%), repFII (60/69, 87.0%), repColE (48/69, 69.6%), repK (43/69, 62.3%) and repI1 (28/69, 41.0%). Other replicons (repP, repFIA, repY, repN, repL/M and repHI2) were detected in 2.9% to 17.4% of the isolates (Figure 2). Among the 45 Ec-MRnoB, 4 major replicon types were detected: repFIB (42/45, 93.3%), repFII (28/45, 62.2%), repFIA (24/45, 53.3%) and repColE (23/45, 51.0%). Furthermore, positive results were detected in some of these isolates for other replicons, including repI1, repY, repP, repB/O, repA/C, repR and repK (Figure 2).
Figure 2.
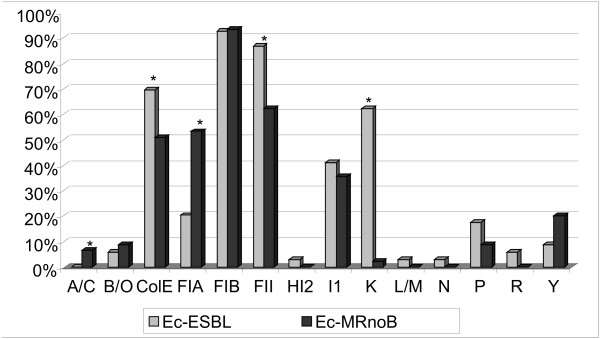
Distribution of replicons on plasmids identified in multiresistant E. coli isolates producing ESBL (Ec-ESBL, n=69) or lacking these enzymes (Ec-MRnoB, n=45). *p value <0.05.
Ec-ESBL and Ec-MRnoB isolates differed significantly in the presence of 5 replicons: repK (p<0.001), repFII (p=0.002) and repColE (p<0.001) were more frequent among Ec-ESBL isolates, while repFIA (p<0.001) and repA/C (p=0.030) were more frequent among Ec-MRnoB isolates.
Nineteen ESBL-producing transconjugants were obtained from the 20 Ec-ESBL isolates selected for the conjugations assays (see Additional file 1). All transconjugants contained one or more plasmids. The more frequently detected replicons in the 19 transconjugants were: repK (14/19, 73.7%), repFII (11/19, 57.9%), repI1 (5/19, 26.3%) and repP (2/19, 10.5%). With the three selective media used, transconjugants were obtained from 13 out of the 20 Ec-MRnoB isolates selected for conjugation assays (see Additional file 2). In all, 25 transconjugants were analysed, including 12 selected with ampicillin, which contained replicons repI1 (n=6), repFIB (n=5), repFII (n=4) and repFIA (n=3), 10 selected with gentamicin [containing plasmids with replicons repFIB (n=6), repFIA (n=4) and repFII (n=4)] and three selected with sulfamethoxazole, [with plasmids containing repFIB (n=2), repFIA (n=2) and repFII (n=2) replicons].
Detection of resistance genes in wild-type strains and transconjugants
In the 69 Ec-ESBL isolates, genes coding for the following ESBL were detected: blaCTX-M-14 (51/69, 73.9%), blaSHV-12 (11/69, 15.9%), blaCTX-M-1 (6/69, 8.7%), blaSHV-2 (5/69, 7.2%), blaCTX-M-9 (2/69, 2.9%) and blaTEM-200 (1/69, 1.4%, first identified in this study). In addition, TEM-1 was detected in 33 (47.8%) isolates. No positive PCR results were obtained in this collection for genes coding for plasmid-mediated AmpC-β-lactamases. The ESBL genes produced by the 19 obtained transconjugants were: blaCTX-M-14 (16/19, 84.2%), blaCTX-M-9 (1/19, 5.3%), blaSHV-12 (1/19, 5.3%) and blaTEM-200 (1/19, 5.3%). Hybridization assays showed that the gene coding for CTX-M-14 was mobilizable by IncK plasmids. Moreover, the RFLP of plasmids from 13 IncK-CTX-M-14 showed the same restriction pattern in 8 isolates (Figure 3) (unfortunately, for the remaining 5 isolates the assay did not allow the production of a restriction pattern).
Figure 3.
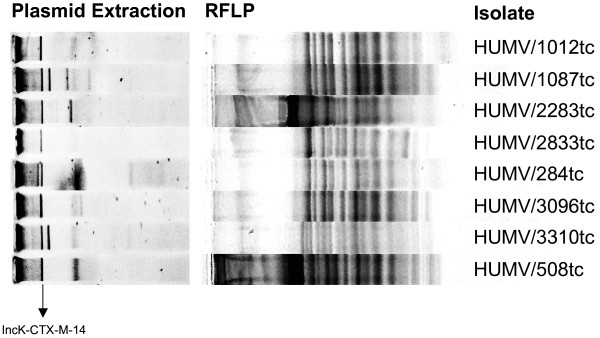
Restriction patterns to the IncK-CTX-M-14-plasmids belong to transconjugants obtained from the ESBL collection.
Amoxicillin resistance in the 45 Ec-MRnoB isolates was related to genes coding for TEM-1 (36/45, 80%), SHV-11 (2/45, 4.4%), SHV-1 (1/45, 2.2%) or OXA-1 (1/45, 2.2%). The relationship between amoxicillin resistance and TEM-1 production in these organisms was confirmed by detecting the corresponding gene in 24 out the 25 (96%) obtained transconjugants. The Ec-MRnoB resistant to both extended-spectrum cephalosporins and cefoxitin contained the gene coding for CMY-2, which was included in IncA/C plasmids, as confirmed by hybridization assays.
None of the 69 Ec-ESBL or the 45 Ec-MRnoB contained any of the studied plasmid-mediated quinolone resistance genes.
In the 69 Ec-ESBL isolates, class 1, class 2 or class 1 plus class 2 integrons were detected in 33.3%, 10.1% and 2.9% isolates, respectively (Table 3). Similarly, for the 45 Ec-MRnoB, positive results for class 1, class 2, and class 1 plus class 2 integrons were obtained for 75.6%, 4.4%, and 6.7% of the isolates (Table 4). The gene cassette arrays found in class 1 integrons for both E. coli collections are shown in Tables 3 and 4.
Table 3.
Distribution of gene cassette arrays found in class 1 integrons among phylogenetic groups of E. coli belongs to Ec-ESBL collection
| |
|
Phylogenetic groups (number of isolates) |
|||
|---|---|---|---|---|---|
| Cassettes | Total | A (N=23) | B1 (N=26) | B2 (N=5) | D (N=15) |
|
ant(3″)-Ia |
3 |
2 (8.7%) |
0 |
0 |
1 (6.7%) |
|
dfrA1-ant(3″)-Ia |
7 |
4 (17.4%) |
2 (7.7%) |
0 |
1 (6.7%) |
|
blaOXA-1-ant(3″)-Ia |
1 |
1 (4.3%) |
0 |
0 |
0 |
|
dfrA12-ant(3″)-Ib |
0 |
0 |
0 |
0 |
0 |
|
dfrA16-ant(3″)-Ib |
4 |
2 (8.7%) |
1 (3.8%) |
0 |
1 (6.7%) |
|
dfrA17-ant(3″)-Ie |
8 |
3 (13%) |
1 (3.8%) |
1 (20%) |
3 (20%) |
|
ant(2″)-Ia |
0 |
0 |
0 |
0 |
0 |
| ant(3″)-Ia-ant(2″)-Ia | 0 | 0 | 0 | 0 | 0 |
Table 4.
Distribution of gene cassette arrays found in class 1 integrons among phylogenetic groups of E. coli belongs to Ec-MRnoB collection
| |
|
Phylogenetic groups (number of isolates) |
|||
|---|---|---|---|---|---|
| Cassettes | Total | A (N=14) | B1 (N=9) | B2 (N=7) | D (N=15) |
|
ant(3″)-Ia |
5 |
0 |
3 (33.3%) |
1 (14.3%) |
1 (6.7%) |
|
dfrA1-ant(3″)-Ia |
6 |
3 (21.4%) |
1 (11.1%) |
0 |
2 (13.3%) |
|
blaOXA-1-ant(3″)-Ia |
1 |
1 (7.1%) |
0 |
0 |
0 |
|
dfrA12-ant(3″)-Ib |
3 |
1 (7.1%) |
0 |
1 (14.3%) |
1 (6.7%) |
|
dfrA16-ant(3″)-Ib |
0 |
0 |
0 |
0 |
0 |
|
dfrA17-ant(3″)-Ie |
16 |
4 (28.6%) |
2 (22.2%) |
4 (57.1%) |
6 (40%) |
|
ant(2″)-Ia |
1 |
0 |
0 |
0 |
1 (6.7%) |
| ant(3″)-Ia-ant(2″)-Ia | 2 | 2 (14.3%) | 0 | 0 | 0 |
Discussion
This study presents comparative information about the microbiological characteristics of two groups of multiresistant clinical isolates of E. coli (producing or not producing ESBL, respectively), recovered in the same geographical and temporal context.
Analysis of Rep-PCR shows a wide clonal distribution among Ec-ESBL isolates and to a lesser extent among Ec-MRnoB isolates. This variability indicates that, in our area, multiresistance in E. coli is not always caused by the expansion of only one or a few clones, but it is often caused by the presence of multiple independent strains. The diversity of E. coli strains producing extended-spectrum beta-lactamase has been previously reported in a nationwide study in Spain [18]. In addition, MLST also showed evidences of small clusters of strains belonging to clonal complexes 354, 10 and 23 or to the sequence types 131, 224, 648 and 117. All these clonal groups have been previously described [19-21] as involved in the spread of certain genes coding for ESBLs and other resistance mechanisms.
Isolates belonging to the ST354Cplx have been related worldwide to the spread of ESBLs of the CTX-M family, associated with the presence of plasmids of different incompatibility groups [19,22]. In Spain, Mora et al. [19] have reported an increased prevalence of strains of ST354 producing CTX-M-14. However, in our study, the ST354 isolates do not produce an ESBL.
The ESBL-producing isolates of the ST10Cplx contained either IncK or IncI1 plasmids, as also described by other authors [23]. IncI1 plasmids have previously been identified in strains of human origin (both in patients and carriers) and in the commensal bacterial flora of diseased animals [24]. ST10Cplx isolates were also identified among non-ESBL detected in our study, but they did not contain IncI1 plasmids.
It has been previously demonstrated that E. coli O25:H4-ST131 is associated to the pandemic dissemination of the CTX-M-15 enzyme but this clone was also prevalent in healthy subjects from different European countries [1]. In a recent study on 100 consecutive extraintestinal E. coli isolates cultured in 2009, the ST131 clone represented 9% of all E. coli and about 25% of all multiresistant isolates in our centre [25]. In the current study, ST131 strains were also identified in both Ec-ESBL and Ec-MRnoB isolates.
CTX-M-14 was the most frequent ESBL identified in our Ec-ESBL isolates. In most cases the gene coding for this enzyme was in IncK plasmids and less frequently in an IncI1 plasmid, in agreement with a previous Spanish report [23]. Moreover, the IncK plasmids identified in this study showed identical restriction patterns (Figure 3), which suggest that the transmission of CTX-M-14 in our sanitary area is due to a specific plasmid belonging to this incompatibility group. Other reports have also shown the importance of CTX-M-14 in other countries, that was disseminated associated to IncFII plasmids [26,27].
The detection of both IncK and IncI1 plasmids in the Ec-MRnoB collection indicates that these mobile elements are not only important for ESBL dispersion, but may also be relevant for the transmission of other resistance mechanism, as suggested in previous reports [7]. On the other hand, resistance to expanded-spectrum cephalosporins associated to the production of the cephamycinase CMY-2 in the Ec-MRnoB was related to a different group of plasmids, namely those of the IncA/C group. IncA/C plasmids coding for CMY-2 have also been previously described in E. coli and Salmonella enterica isolates [7]. Moreover, 4 isolates were resistant to ceftazidime but they did not present plasmid-mediated AmpC β-lactamases, we presume that hyperproduction of AmpC was due to mutation in the promotor or the attenuator of the corresponding gene, as observed previously by others authors [28].
Plasmid typing showed that the dichotomous distribution of CTX-M-14 and CMY-2 among the two E. coli groups corresponded to an unequal distribution of two plasmid types associated to these enzymes: the A/C plasmids carrying CMY-2 were unique to the EcMRnoB group, while the IncK plasmids carrying CTX-M-14 were related to the Ec-ESBL group.
Interestingly, other plasmid species were common and highly represented in the two groups of isolates: IncF, ColE and IncI1. The high prevalence of IncF plasmids in both Ec-ESBL and Ec-MRnoB clearly indicates that this plasmid species is very well adapted in resistant E. coli strains independently of their resistance phenotype. A recent report has demonstrated that F replicons (FIA, FIB, FIC and FII) were the most frequently detected replicon types in E. coli strains producing or not producing ESBL [29]. Replicons of the IncF type were detected in 50% of E. coli from faeces of healthy, antibiotic-free humans and faecal flora from healthy birds in the USA, confirming that this plasmid type can be highly represented in E. coli populations also including susceptible strains [24]. Similarly, IncI1 plasmids have also been detected in E. coli from faecal flora of healthy humans and animals [24]. Finally, ColE plasmids are small, high copy number, not self-conjugative, producing bacteriocins, whose prevalence is not well estimated in recent collections of Enterobacteriaceae.
Several studies [30,31] have indicated that extraintestinal E. coli isolates are more commonly of phylogenetic groups B2 and D than of groups A and B1. In our series, groups D and B2 were more frequent in the Ec-MRnoB collection than in the Ec-ESBL. The decreased level of resistance among isolates of group B2 reported in some studies [32] was not observed in our case, as per definition all isolates were multiresistant. Although multiple studies indicate that use of fluoroquinolones is an independent factor for infections by multiresistant E. coli, plasmid-mediated quinolone resistance genes were not found among the isolates we have studied. The most likely explanation for quinolone resistance in our isolates is the presence of chromosomal mutations in their gyrase and topoisomerase IV genes.
In addition to the indicated resistance genes, we have found that the clinical isolates of multiresistant E. coli in our health area carry different classes of integrons. Ec-MRnoB showed a higher presence of these elements in comparison with the isolates belonging to the Ec-ESBL collection but, in both cases, the class 1 integrons containing dfrA17-ant(3′)Ie or dfrA1-ant(3″)-Ia genes were the most frequent ones. The implication of these elements in the spread of resistance in Spain [33] has been previously documented.
Conclusion
In conclusion, this study has shown that, in our area, multiresistant E. coli producing either ESBL or other mechanisms of resistance are clonally diverse, although small clusters of related strains are also found. While both Ec-ESBL and EcMRnoB frequently contained IncFI plasmids, plasmids usually related to the most frequently detected ESBL (CTX-M-14), are uncommonly found in strains lacking this enzyme.
Methods
Bacterial isolates, susceptibility testing and clonal relationship
Two hundred multiresistant E. coli (one per patient) producing (n=100) or not producing ESBL (n=100), consecutively obtained between January 2004 and February 2005 at the Clinical Microbiology Service of the University Hospital Marqués de Valdecilla (Santander, Spain) were initially considered for this study. The organisms were obtained from urine (n=158) or from other samples (n=42, including 17 wound exudates, 8 samples from blood, 6 sputum, 6 naso-pharyngeal lavage, 2 catheter, 2 ascitic liquid and 1 bronchoalveolar aspirate). One hundred and sixteen isolates were from samples of patients admitted to the hospital and 84 from outpatients (database from Hospital Universitario Marqués de Valdecilla). No relevant differences were observed in the distribution of these parameters when comparing Ec-ESBL and Ec-MRnoB.
Identification and preliminary susceptibility testing (including ESBL production) of the isolates had been routinely performed with the WalkAway system (Dade Behring, Inc., West Sacramento, Ca., USA) using gram-negative MIC combo 1S panels. Confirmation of ESBL production and determination of MICs of imipenem, meropenem, aztreonam, piperacillin, cefoxitin, cefotetan, cefotaxime, cefotaxime-clavulanic acid, ceftazidime, ceftazidime-clavulanic acid and cefepime were performed using Dried MicroScan ESβL plus (Dade Behring, Inc., West Sacramento, Calif.) panels according to the manufacturer’s recommendations. In addition, susceptibility testing results of the following agents were confirmed by standardized microdilution, according to the CLSI standard [18]: amoxicillin (Sigma-Aldrich, Madrid, Spain), amoxicillin-clavulanic acid (GlaxoSmithKline, United Kingdom) (2:1), cefepime-clavulanic acid, piperacillin (Sigma-Aldrich)-tazobactam (Wyeth Pharmaceuticals, Madrid, Spain), amikacin (Sigma-Aldrich), gentamicin (Sigma-Aldrich), tobramycin (Sigma-Aldrich), nalidixic acid (Sigma-Aldrich), ciprofloxacin (Fluka, Madrid, Spain), tetracycline (Sigma-Aldrich), doxycycline (Sigma-Aldrich), and trimethoprim (Sigma-Aldrich)-sulfamethoxazole (Sigma-Aldrich) (19:1). Fixed concentrations of 4 mg/L of tazobactam or clavulanic acid were used in combination with piperacillin and cefepime, respectively. The results were interpreted according to the EUCAST breakpoints [17]. Isolates lacking ESBL were selected for this study if resistant to at least three of the following agents: amoxicillin, amoxicillin-clavulanic acid, nalidixic acid, gentamicin or tobramycin and trimethoprim-sulfamethoxazole. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as control strains in susceptibility testing assays. Phylogenetic grouping of the 200 isolates was determined by multiplex PCR, as described by Clermont et al. [19].
Clonal relationship was determined by Rep-PCR as previously described [20]. Amplicons were run in a 1.5% agarose gel for 100 min, stained with ethidium bromide (Sigma Chemical CO. St. Louis, USA) and photographed. Two isolates were considered to be clonally unrelated when at least two different bands were observed. Clonal relationship among isolates was also determined by XbaI-PFGE [21] when ESBL-producing isolates showed the same Rep-PCR pattern than isolates lacking ESBL, these isolates were also analysed by MLST, and this assay was also performed for 40 isolates selected for the conjugation assay representing the most frequent Rep-PCR patterns of each E. coli collection (see below). Detection by PCR and sequencing of 7 housekeeping genes (gyrB, adk, purA, recA, icd, mdh and fumC) were performed according to the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli).
Plasmid profile and hybridization experiments
After observing that some isolates with the same Rep-PCR pattern presented different clinical categories of at least two antimicrobial agents of different classes, 69 Ec-ESBL isolates and 45 Ec-MRnoB isolates were selected for plasmid analysis and detection of plasmid-mediated genes coding for resistance to β-lactams and quinolones, according to the following criteria: a) all isolates from each Rep-PCR pattern with single isolates, b) one isolate of each antimicrobial susceptibility pattern from Rep-PCR patterns containing >=2 isolates.
Plasmid DNA was extracted by the Kado-Liu method [22] and separated on 0.9% horizontal agarose gels electrophoresis. Plasmids R27 (169 kb, Genbank access AF250878), R1 (94 kb, Genbank access NC_003277), RP4 (55 kb, [23]), and ColE1 (6 kb, Genbank access J01566) were used as size standards.
Plasmids were also characterized by PCR-based replicon typing (PBRT), as described elsewhere, using the respective PBRT controls [4,5]. The obtained amplicons were sequenced to confirm their identity.
Plasmids were transferred onto nylon membranes by southern blotting (Roche, Mannheim, Germany). Amplicons from blaCTX-M-14 and blaCMY-2 were obtained by PCR using DNA from E. coli CCG02 and E. coli B-12 [24], respectively. Similarly, plasmids R387 and pIP40a [5] were used to obtain PCR amplicons from repK and repA/C, respectively. DNA probes prepared with DIG-High Prime (Roche, Penzberg, Germany) were used to investigate the presence of blaCTX-M-14 and repK genes in the same plasmid of Ec-ESBL isolates and of blaCMY-2 and repA/C genes in the same plasmid of Ec-MRnoB isolates.
In 13 transconjugants of the belonging to ESBL collection the relationship among repK-CTX-M-14-plasmids was determined by comparison of their DNA patterns generated after digestion with the EcoRI and PstI enzymes and electrophoresis in 1.5% agarose, as described elsewhere [25].
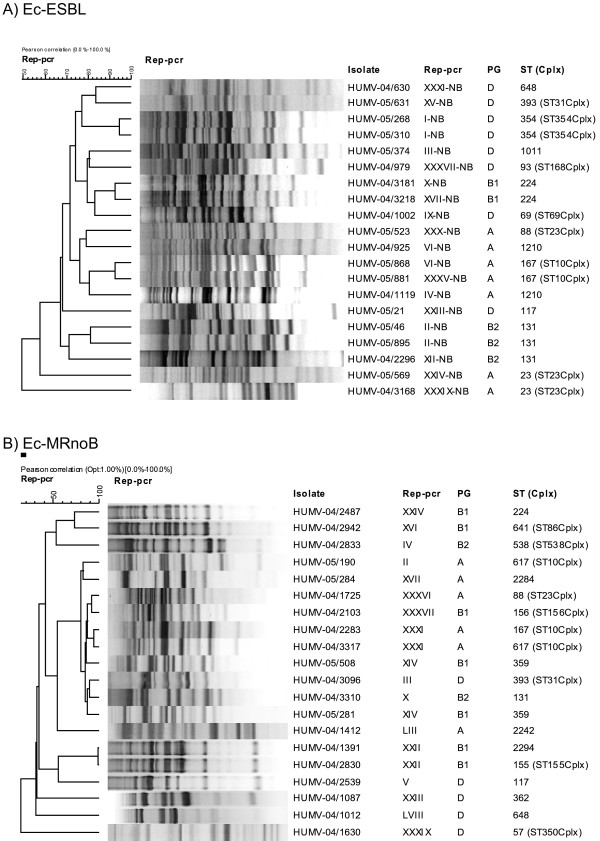
Conjugation assays
Conjugation assays were performed with 20 Ec-ESBL and 20 Ec-MRnoB, which are representative of the most common Rep-PCR/antibiotic resistance patterns (Figure 4). E. coli J53 resistant to sodium azide was used as a recipient strain. Transconjugants from the Ec-ESBL isolates were selected with sodium azide (100 mg/L) plus cefotaxime (2 mg/L), while for the Ec-MRnoB, transconjugants were selected on three different media: sodium azide (100 mg/L) plus ampicillin (100 mg/L), gentamicin (8 mg/L) or sulfamethoxazole (1000 mg/L).
Figure 4.
Clonal relationship between isolates selected for conjugation assays in both E. coli collections. A) Ec-ESBL, B) Ec-MrnoB.
Detection of resistance determinants
Five multiplex PCRs (Table 5) were performed using previously published conditions to detect genes that are usually included in conjugative plasmids: blaTEM, blaSHV, blaOXA-1 and blaPSE-1[26], plasmid-mediated AmpC-type enzymes [27], blaCTX-M β-lactamases [26], plasmid-mediated quinolone-resistance genes, including qnrA, qnrB, qnrS, aac(6′)-Ib-cr and qepA[28] and tetracyclines-resistance genes tet(A), tet(B) and tet(G) [26]. The identity of the complete genes detected by the multiplex PCR was confirmed by specific PCR (using appropriate primers) and sequencing of the two DNA strands. Finally, class 1 and class 2 integrons were detected by PCR (Table 5) and the variable regions of class 1 integrons were sequenced using specific primers for the 3′CS and 5′CS ends as described elsewhere [29].
Table 5.
PCR conditions used for detection of antimicrobial resistance genes
| Resistance | Gene | Primers | Tm (°C) | Amplicon size (pb) |
|---|---|---|---|---|
| β-Lactams |
blaTEM-1 |
5′-ttgggtgcacgagtgggt-3′ |
60 |
503 |
| 5′-taattgttgccgggaagc-3′ | ||||
|
blaoxa-1 |
5′-agcagcgccagtgcatca-3′ |
708 |
||
| 5′-attcgaccccaagtttcc-3′ | ||||
|
blapse-1 |
5′-cgcttcccgttaacaagtac-3′ |
419 |
||
| 5′-ctggttcatttcagatagcg-3′ | ||||
|
blashv |
5′-tcgggccgcgtaggcatg-3′ |
606 |
||
| 5′-agcagggcgacaatcccg-3′ | ||||
| Quinolones |
qnrA |
5′-agaggatttctcacgccagg-3′ |
56 |
579 |
| 5′-tgccaggcacagatcttgac-3′ | ||||
|
qnrB |
5′-ggmathgaaattcgccactg-3′ |
263 |
||
| 5′-ttygcbgyycgccagtcggcg-3′ | ||||
|
qnrS |
5′-gcaagttcattgaacagggt-3′ |
427 |
||
| 5′-tctaaaccgtcgagttcggcg-3′ | ||||
|
qepA |
5′-aactgcttgagcccgtagat-3′ |
189 |
||
| 5′-cgtgttgctggagttcttcc-3′ | ||||
|
aac(6′)-Ib-cr |
5′-ttgcaatgctgaatggagag-3′ |
218 |
||
| 5′-cgtttggatcttggtgacct-3′ | ||||
| Cephalosporins |
blactx-m-like |
5′-cgatgtgcagtaccagtaa-3′ |
60 |
585 |
| 5′-ttagtgaccagaatcagcgg-3′ | ||||
|
blactx-m-group1 |
5′-atggttaaaaaatcactgcg-3′ |
876 |
||
| 5′-ttacaaaccgtcggtgac-3′ | ||||
|
blactx-m-group9 |
5′-atggtgacaaagagagtgcaac-3′ |
876 |
||
| 5′-ttacagcccttcggcgatg-3′ | ||||
|
blactx-m-group2 |
5′-tcaagaagagcgacctggtt-3′ |
601 |
||
| 5′-gatacctcgctccatttattg-3′ | ||||
| Cephamycinases |
blaacc |
5′-aacagcctcagcagccggtta-3′ |
64 |
346 |
| 5′-ttcgccgcaatcatccctagc-3′ | ||||
|
blacit |
5′-tggccagaactgacaggcaaa-3′ |
462 |
||
| 5′-tttctcctgaacgtggctggc-3′ | ||||
|
bladha |
5′-aactttcacaggtgtgctgggt-3′ |
405 |
||
| 5′-ccgtacgcatactggctttgc-3′ | ||||
|
blamox |
5′-gctgctcaaggagcacaggat-3′ |
520 |
||
| 5′-cacattgacataggtgtggtg-3′ | ||||
|
blaebc |
5′-tcggtaaagccgatgttgcgg-3′ |
302 |
||
| 5′-cttccactgcggctgccagtt-3′ | ||||
|
blafox |
5′-aacatggggtatcagggagatg-3′ |
190 |
||
| 5′-caaagcgcgtaaccggattgg-3′ | ||||
| Tetracyclines |
tet (G) |
5′-gctcggtggtatctctgc-3′ |
60 |
500 |
| 5′-agcaacagaatcgggaac-3′ | ||||
|
tet (A) |
5′-gctacatcctgcttgcct-3′ |
210 |
||
| 5′-catagatcgccgtgaaga-3′ | ||||
|
tet (B) |
5′-ttggttaggggcaagttttg-3′ |
600 |
||
| 5′-gtaatgggccaataacaccg-3′ | ||||
| Integrons |
intI1 |
5′-gccttgctgttcttctac-3′ |
55 |
558 |
| 5′-gatgcctgcttgttctac-3′ | ||||
|
intI2 |
5′-cacggatatgcgacaaaaaggt-3′ |
65 |
788 |
|
| 5′-gtagcaaacgagtgacgaaatg-3′ | ||||
| Int-class 1 (IC1) |
5′-ggcatccaagcagcaagc-3′ |
58 |
variable |
|
| 5′-aagcagacttgacctgat-3′ | ||||
| Int-class 2 (IC2) |
5′-cgggatcccggacggcatgcacgatttgta-3′ |
65 |
variable |
|
| 5′-gatgccatcgcaagtacgag-3′ |
Statistical methods
The proportions of Ec-ESBL and of Ec-MRnoB isolates containing different replicon elements were compared by the chi-square test. Differences were considered significant for p<0.05. The test was performed using the SPSS 11.0 program.
Competing interest
Luis Martínez.-Martínez: reports that he has been a consultant for Wyeth and Pfizer, has served as speaker for Wyeth, Merck, Pfizer, and Janssen-Cilag, and has received research support from Merck, Wyeth, and Janssen-Cilag. The other authors declare that they have no competing interests.
Authors’ contributions
BRdC carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. EJR carried out the molecular genetic studies. LV and CT participated in the design of the study and performed the statistical analysis. BG, AC and LMM conceived the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Resistance phenotype and resistance genes in 19 EcESBL and their derived transconjugants.
Resistance phenotype and resistance genes in 13 Ec-MRnoB and their derived transconjugants.
Contributor Information
Belén Ruiz del Castillo, Email: biolbelen@hotmail.com.
Laura Vinué, Email: lauravinue@hotmail.com.
Elena Jesús Román, Email: nena_roman@hotmail.com.
Beatriz Guerra, Email: beatriz.guerra@bfr.bund.de.
Alessandra Carattoli, Email: alecara@iss.it.
Carmen Torres, Email: carmen.torres@unirioja.es.
Luis Martínez-Martínez, Email: lmartinez@humv.es.
Acknowledgements
This study was supported by Ministerio de Economía y Competitividad, Instituto de Salud Carlos III- co-financed by European Development Regional Fund “A way to achieve Europe” ERDF, Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015), by FIS (grant PI050690), the Fundación Leonardo Torres Quevedo, University of Cantabria (grant FMV-UC 2008–01) and the Federal Institute for Risk Assessment (BfR, Berlin; BfR-45-004). B. R. del C. and E.R. are supported by IFIMAV, Cantabria, Spain. B. Ruiz performed a four-month stay at the Department of Biological Safety of the BfR supported by a SEIMC short visit grant. We acknowledge Dr. G. Prats for providing us with the E. coli strains RS218, ECOR39, ECOR16, ECOR1 and ECOR32 used as controls of phylogenetic groups. We acknowledge Sophia Mooney for her review of the English version of the manuscript.
References
- Woodford N. Successful, multiresistant bacterial clones. J Antimicrob Chemother. 2008;61(2):233–234. doi: 10.1093/jac/dkm474. [DOI] [PubMed] [Google Scholar]
- Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing enterobacteriaceae in europe. Euro Surveill. 2008;13(47):pii:19044. [PubMed] [Google Scholar]
- Rodríguez-Baño J, Picón E, Gijón P, Hernández JR, Ruíz M, Peña C, Almela M, Almirante B, Grill F, Colomina J. Community-onset bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli: risk factors and prognosis. Clin Infect Dis. 2010;1(50):40–48. doi: 10.1086/649537. [DOI] [PubMed] [Google Scholar]
- García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother. 2009;63(2):274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- Carattoli A, Bertinia A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33(3):657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A, Canton R, Valverde A, Machado E, Galan JC, Peixe L, Carattoli A, Baquero F, Coque TM. Dissemination and Persistence of blaCTX-M-9 Are Linked to Class 1 Integrons Containing CR1 Associated with Defective Transposon derivatives from Tn402 Located in Early Antibiotic Resistance Plasmids of IncHI2, IncP1, and IncFI Groups. Antimicrob Agents Chemother. 2006;50(8):2741–2750. doi: 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KL, Liebana E, Villa L, Batchelor M, Threlfall EJ, Carattoli A. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob Agents Chemother. 2006;50(9):3203–3206. doi: 10.1128/AAC.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother. 2009;53(10):4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gołebiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, Gniadkowski M, Bardowski J, Cegłowski P. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother. 2007;51(11):3789–3795. doi: 10.1128/AAC.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmin Kim Y-ML, Jeong Y-S, Seol S-Y. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 Extended-Spectrum beta-Lactamases in Enterobacteriaceae Clinical Isolates in Korea. Antimicrob Agents Chemother. 2005;49(4):1572–1575. doi: 10.1128/AAC.49.4.1572-1575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TM Coque AN, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis. 2008;14(2):195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakkoupi P, Vourli S, Vatopoulos AC, Kanellopoulou M, Papafrangas E, Raitsiu B. A multiresistant Klebsiella pneumoniae clinical isolate carrying both CTX-M-15 and VIM-1 beta-lactamases, harboured by different plasmids. Int J Antimicrob Agents. 2009;33(2):191–192. doi: 10.1016/j.ijantimicag.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Diestra K, Juan C, Curiao T, Moya B, Miro E, Oteo J, Coque TM, Perez-Vazquez M, Campos J, Canton R. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother. 2009;63(1):60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- Gołebiewski IK-Z M, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, Gniadkowski M, Bardowski J, Cegłowski P. Complete Nucleotide Sequence of the pCTX-M3 Plasmid and Its Involvement in Spread of the Extended-Spectrum beta-Lactamase Gene blaCTX-M-3. Antimicrob Agents Chemother. 2007;51(11):3789–3795. doi: 10.1128/AAC.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. 2011. Version 3.1, 2013. http://www.eucast.org.
- Díaz MA, Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, Pascual A. for the Spanish Group for Nosocomial Infections (GEIH) Diversity of Escherichia coli strains producing extended-spectrum beta-lactamases in Spain: second nationwide study. J Clin Microbiol. 2010;48(8):2840–2845. doi: 10.1128/JCM.02147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Blanco M, López C, Mamani R, Blanco JE, Alonso MP, García-Garrote F, Dahbi G, Herrera A, Fernández A. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents. 2011;37(1):16–21. doi: 10.1016/j.ijantimicag.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Crémet L, Caroff N, Giraudeau C, Dauvergne S, Lepelletier D, Reynaud A, Corvec S. Occurrence of ST23 complex phylogroup A Escherichia coli isolates producing extended-spectrum AmpC beta-lactamase in a French hospital. Antimicrob Agents Chemother. 2010;54(5):2216–2218. doi: 10.1128/AAC.01580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, El-Defrawy I, Abou-Aitta A, Klena J, Nicolas-Chanoine MH. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microbiology Drug Resistance. 2011;17(1):67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis. 2008;14(2):195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde A, Cantón R, Garcillán-Barcia MP, Novais A, Galán JC, Alvarado A, De la Cruz F, Baquero F, Coque TM. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother. 2009;53(12):5204–5212. doi: 10.1128/AAC.01706-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Applied and Enviromental Microbiology. 2007;73(6):1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, García-Garrote F. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother. 2011;66(9):2011–2021. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- Cao X, Cavaco LM, Lv Y, Li Y, Zheng B, Wang P, Hasman H, Liu Y, FM A. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol. 2011;49(7):2496–2501. doi: 10.1128/JCM.02503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PL, Yeung MK, Lo WU, Tse H, Li Z, Lai EL, Chow KH, To KK, WC Y. Predominance of pHK01-like incompatibility group FII plasmids encoding CTX-M-14 among extended-spectrum beta-lactamase-producing Escherichia coli in Hong Kong, 1996–2008. Diagn Microbiol Infect Dis. 2012;73(2):182–186. doi: 10.1016/j.diagmicrobio.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Ortega A, Oteo J, Aranzamendi-Zaldumbide M, Bartolomé RM, Bou G, Cercenado E, Conejo MC, González-López JJ, Marín M, Martínez-Martínez L. Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother. 2012;56(7):3576–3581. doi: 10.1128/AAC.06393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcade G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J Antimicrob Chemother. 2009;63(1):67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- Moreno E, Prats G, Sabate M, Perez T, Johnson JR, Andreu A. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother. 2006;57:204–211. doi: 10.1093/jac/dki468. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, Paulish TE, Mordechai E, Adelson ME, Gygax SE, Trama JP. Antimicrobial non-susceptibility of cervico-vaginal and rectal Escherichia coli isolates is associated with phylogeny and plasmid carriage. European Journal of Clinical Microbiology and Infections Disseases. 2009;28(11):1399–1403. doi: 10.1007/s10096-009-0788-3. [DOI] [PubMed] [Google Scholar]
- Vinué L, Sáenz Y, Somalo S, Escudero E, Moreno MA, Ruiz-Larrea F, Torres C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J Antimicrob Chemother. 2008;62(5):934–937. doi: 10.1093/jac/dkn331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Resistance phenotype and resistance genes in 19 EcESBL and their derived transconjugants.
Resistance phenotype and resistance genes in 13 Ec-MRnoB and their derived transconjugants.