Significance
Nucleocytoplasmic transport requires the orchestrated assembly and disassembly of cargo–carrier complexes. Visualizing the dynamics of these interactions during nuclear import and export with millisecond time resolution using single-molecule FRET provides new insight into the mechanism of the transport cycle. We show that although key interactions between components are reversible and multiple outcomes are often possible, effective transport is generated through precise spatiotemporal control of cargo–carrier complex assembly and disassembly. The novel transient intermediates identified and the kinetic models developed emphasize that the distinct environments at the nuclear and cytoplasmic faces of nuclear pores are necessary for productive transport.
Keywords: FRET, nucleocytoplasmic transport
Abstract
Nuclear pore complexes (NPCs) mediate the exchange of macromolecules between the cytoplasm and the nucleoplasm. Soluble nuclear transport receptors bind signal-dependent cargos to form transport complexes that diffuse through the NPC and are then disassembled. Although transport receptors enable the NPC's permeability barrier to be overcome, directionality is established by complex assembly and disassembly. Here, we delineate the choreography of importin-α/CAS complex assembly and disassembly in permeabilized cells, using single-molecule fluorescence resonance energy transfer and particle tracking. Monitoring interaction sequences in intact NPCs ensures spatiotemporal preservation of structures and interactions critical for activity in vivo. We show that key interactions between components are reversible, multiple outcomes are often possible, and the assembly and disassembly of complexes are precisely controlled to occur at the appropriate place and time. Importin-α mutants that impair interactions during nuclear import were used together with cytoplasmic Ran GTPase-activating factors to demonstrate that importin-α/CAS complexes form in the nuclear basket region, at the termination of protein import, and disassembly of importin-α/CAS complexes after export occurs in the cytoplasmic filament region of the NPC. Mathematical models derived from our data emphasize the intimate connection between transport and the coordinated assembly and disassembly of importin-α/CAS complexes for generating productive transport cycles.
Nuclear pore complexes mediate the transport of macromolecules across the double-membrane nuclear envelope (NE) of eukaryotic cells. Each nuclear pore complex (NPC) is estimated to transport in excess of 10 MDa of material, or more than 30 molecules, each second (1, 2). Signal-dependent transport through NPCs is mediated by soluble transport receptors that bind their cargo in the donor compartment and release it in the target compartment after diffusing through an NPC. However, detailed analysis of this process is complicated by the bidirectional nature of cargo movement within NPCs and by the assembly and disassembly of cargo complexes occurring concomitantly with translocation. Metazoan NPCs have a cylindrical central transport channel ∼90 nm long and ∼50 nm in diameter from which flexible filaments extend outward (up to ∼200 nm) into the cytoplasm (3). Filaments (∼75 nm) also extend into the nucleoplasm, but terminate at the distal ring, together forming the nuclear basket (4, 5). Despite their immense size (∼60–120 MDa), NPCs are constructed from only ∼30 different proteins [nucleoporins (Nups)], each of which is present in an integer multiple of eight copies (4, 6, 7).
Mechanistic models of nuclear transport seek to explain how NPCs show such remarkable selectivity, allowing only a defined subset of molecules to permeate through them. The answer lies in understanding the properties of the material that occupies the central pore. This channel is occupied by natively unfolded proteins containing, in aggregate, thousands of phenylalanine–glycine (FG) repeat motifs (7–11). In the absence of other factors, the log of the rate constant to penetrate this barrier is approximately linearly dependent on the Stokes radius of the particle (12). However, the surface properties of the particle are also important, as perturbations to hydrophobicity or charge can significantly alter permeability rates (13, 14). Large molecules that, based on size alone, should have low permeabilities can have very rapid translocation rates if they can also interact with the FG motifs (e.g., transport receptors). In general, small molecules (< ∼20–40 kDa) do not need to be recognized specifically and simply diffuse through NPCs (for reviews, see refs. 15 and 16).
Transport receptors of the importin-β (Imp-β) superfamily recognize signal sequences on cargo molecules and also interact with FG motifs, thereby promoting movement of cargo through NPCs (15–17). The importin-α (Imp-α) adaptor increases the range of cargos that can be transported by Imp-β. After transport into the nucleus, the heterotrimeric import complex consisting of Imp-β, Imp-α, and cargo is dissociated by the GTP-bound form of the G protein, Ran (18). With the exception of mRNA carriers, export complexes invariably contain RanGTP. For example, both Imp-β and Imp-α return to the cytoplasm with RanGTP, although Imp-α additionally requires its specific transport factor, CAS. Cytoplasmically located Ran binding proteins (RanBPs) and RanGAP activate Ran’s GTPase, freeing Imp-β and Imp-α for another round of cargo transport (19–21) (reviewed by refs. 15–17).
Overall transport directionality is governed by the Ran nucleotide state, which controls both the assembly and the disassembly of transport complexes. A high nuclear RanGTP concentration and low cytoplasmic RanGTP concentration drive both import and export. In principle, translocation through the NPC and the assembly and disassembly of transport complexes can be disconnected processes, although greater transport efficiencies are expected if these processes are coupled (22). Numerous studies have indicated that proteins found on the nucleoplasmic and cytoplasmic faces of NPCs promote key assembly and disassembly steps at the beginning or end of cargo transport (for reviews, see refs. 22–24). However, because transport occurs in the millisecond time regime, it is unclear to what extent these in vitro biochemical results relate to the rapid kinetics that occur during transport through intact NPCs. For example, although the nuclear basket nucleoporin Nup50 (Nup2 in yeast) catalyzes the dissociation of Imp-α/cargo complexes in vitro (25–29), CAS and RanGTP are also required for this reaction during real-time transport through intact NPCs (30).
To establish a more precise spatial and temporal picture of the formation and dissolution of the Imp-α/CAS complex, we used single-molecule fluorescence resonance energy transfer (smFRET) to monitor the partner molecules that are bound to Imp-α during both nuclear import and export in permeabilized cells. We used a range of Imp-α mutants that impair interactions with cargo, Nup50, or CAS, together with the addition of RanGAP and RanBP1 to delineate the importance of complex assembly and disassembly at the nuclear and cytoplasmic faces of NPCs in generating productive nuclear protein import cycles. These approaches address different questions from those previously investigated biochemically, and the mechanistic features revealed using smFRET underline the importance of real-time measurements in functional NPCs for establishing the molecular mechanisms of nucleocytoplasmic transport.
Results and Discussion
Validation of the smFRET Approach.
We previously validated the smFRET approach for real-time measurements of nucleocytoplasmic transport (30). In short, positions of single molecules are detected by narrow-field epifluorescence microscopy and the oligomerization state of the donor-dye tagged protein vis-à-vis the acceptor-tagged protein is determined by FRET. The four solvent-accessible cysteines of human Imp-α were tagged with Alexa568 (donor dye). Wild-type CAS also has four solvent-accessible cysteines, but, when these cysteines were tagged with Alexa647 (acceptor dye), the decrease in the donor-dye emission signal due to FRET was only ∼8%. Therefore, we engineered a CAS mutant (muCAS) containing two additional cysteines (N136C + N186C), giving a total of six. FRET between fully labeled Alexa568-Imp-α and Alexa647-muCAS was ∼41 ± 13% (Fig. S1). The basic tenet of our approach is that FRET is observed only when Imp-α and CAS are bound together in a complex, whereas FRET is lost when they dissociate. Using FRET, the dissociation constant for the Alexa568-Imp-α/Alexa647-muCAS complex was determined as 12 ± 1 nM (Fig. S2), comparable to that determined biochemically (21). In a bulk export assay, muCAS was indistinguishable from wild-type CAS (Fig. S1D). Using these fluorescent Imp-α and CAS constructs, we were able to detect formation of the Imp-α/CAS complex during import of the model cargo NLS-2xGFP(4C) [which contains a nuclear localization signal (NLS) and two green fluorescence protein (GFP) domains] (31) and the dissociation of the Imp-α/CAS complex during export (Fig. 1 and Movies S1 and S2). In these experiments, we excited the donor dye (at 561 or 568 nm) and simultaneously collected the donor and acceptor fluorescence emissions on opposite halves of the same camera with 1-ms time resolution. The protein with the donor dyes (typically Imp-α) was present at ∼0.1 nM, and the protein with acceptor dyes (CAS or the cargo) was present at 250 nM. Because only donor excitation was used, the protein with the acceptor dyes was undetectable unless in close proximity to the donor as part of a complex.
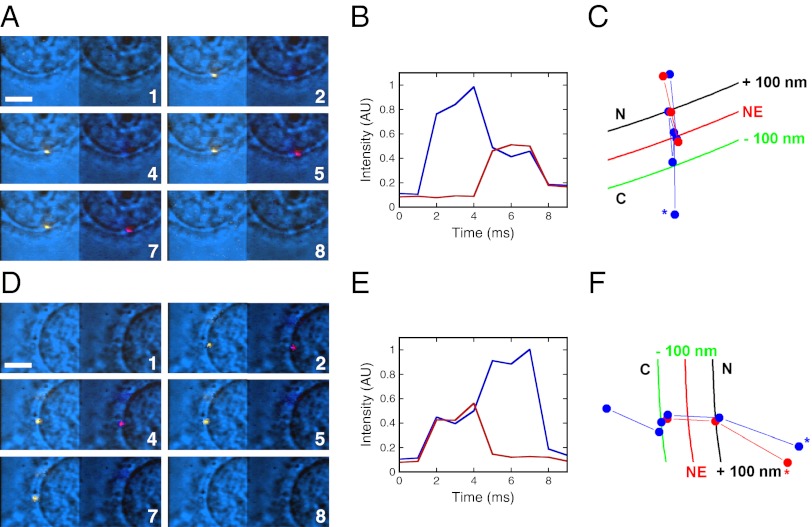
Fig. 1.
Interaction between Imp-α and muCAS detected by smFRET during nuclear import and export. (A) Movie frames showing the appearance of FRET between Alexa568-Imp-α (donor fluorescence is yellow, Left) and Alexa647-muCAS (acceptor fluorescence is red, Right), indicating that an Imp-α/CAS complex was formed during nuclear import. The bright-field background image underlay (blue) shows the position of the NE as a centrally located curve that bisects the images. Numbers denote time (milliseconds). [muCAS] = 250 nM, [Imp-α] = 0.1 nM, [NLS-2xGFP(4C)] = 250 nM, [Imp-β] = 0.5 μM, [Ran] = 2 μM, [NTF2] = 1 μM, and [GTP] = 1 mM. Excitation wavelength: 568 nm. (Scale bar: 5 μm.) (B) Intensity time traces for the interaction event shown in A. Blue, donor fluorescence; red, acceptor fluorescence. (C) Particle trajectories for the interaction event shown in A. The 200-nm distance between the black and green curves approximates the span of the NPC. The asterisk identifies the beginning of the Imp-α trajectory. C, cytoplasm; N, nucleoplasm. Blue, Imp-α; red, muCAS. (D–F) Similar to A–C, but showing the disappearance of FRET between Alexa568-Imp-α and Alexa647-muCAS, indicating that an Imp-α/CAS complex dissociated during nuclear export. The export assay is described in Materials and Methods. [muCAS] = 250 nM, [Ran] = 2 μM, [NTF2] = 1 μM, and [GTP] = 1 mM.
The locations at which Imp-α/CAS complexes assembled and disassembled were determined from the simultaneous particle position and smFRET measurements (Fig. 1). We took that Imp-α/CAS complex formation occurred near the first location at which smFRET was observed during import and that Imp-α/CAS complex dissociation occurred near the last location at which smFRET occurred during export (Fig. 2). These data are consistent with the model that Imp-α/CAS complexes form in the nuclear basket region and disassemble in the cytoplasmic filament region and are consistent with the known positions of Nup50 and RanBP2-RanGAP that in vitro catalyze Imp-α/CAS assembly and disassembly, respectively. Nup50 promotes Imp-α/CAS complex formation during nuclear import (26) and is located primarily at the NPC nuclear face. RanGAP, together with a RanBP, promotes GTP hydrolysis that leads to disassembly of the Imp-α/CAS/RanGTP complex (reviewed by ref. 15) and is located primarily on the filaments that protrude from the cytoplasmic face. The Imp-α/CAS complex formation position (66 ± 44 nm from the midline of the NE) is consistent with the Imp-α/cargo complex dissociation position (87 ± 38 nm) determined previously (30). Imp-α/CAS complex assembly and disassembly occur at the same location for abortive and complete transport events (Fig. 2).
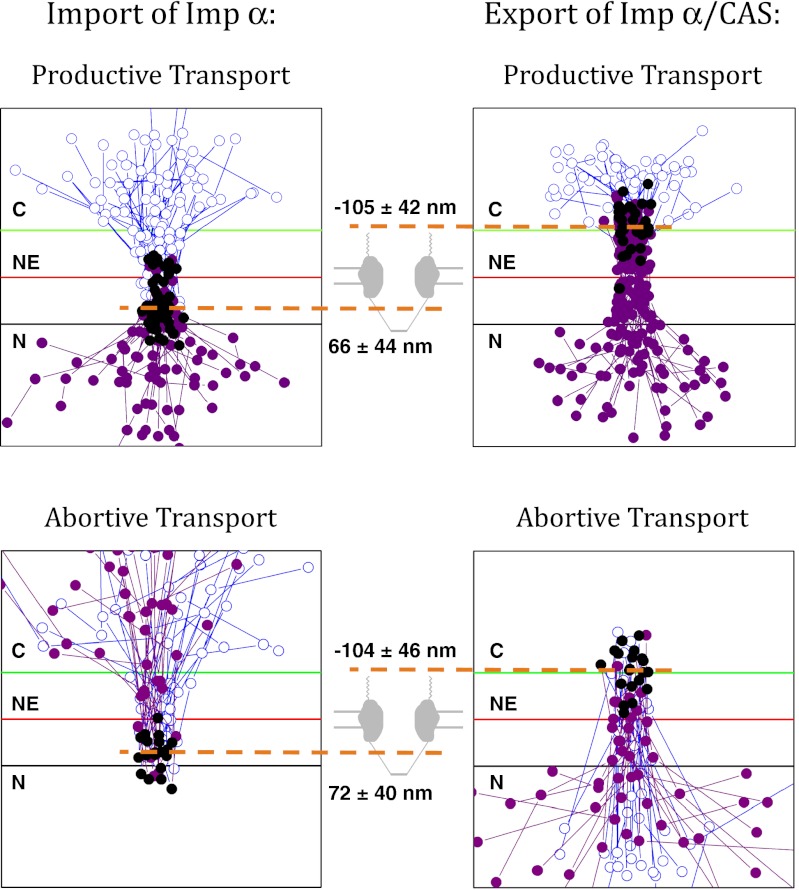
Fig. 2.
Locations at which Imp-α/CAS complexes assemble and disassemble during nuclear import (Left) and export (Right). (Left) Trajectories from nuclear import assays (as in Fig. 1 A–C) in which Imp-α (donor dyes) interacted with CAS (acceptor dyes) and did not dissociate from CAS. (Upper) Those trajectories in which Imp-α ended up in the nucleus (productive transport); (Lower) those trajectories in which Imp-α ended up returning back to the cytoplasm (abortive transport). The positions of CAS-free Imp-α are indicated by blue open circles, and the positions of Imp-α/CAS complexes, as determined by smFRET, are identified by purple solid circles. The black solid circles identify the first location at which FRET was observed (n = 50 and 20, Upper and Lower, respectively). (Right) Trajectories from nuclear export assays of Imp-α/CAS complexes (as in Fig. 1 D–F) in which the Imp-α/CAS complexes dissociated at the NPC. (Upper) Those trajectories in which Imp-α ended up in the cytoplasm (productive transport); (Lower) those trajectories in which Imp-α ended up returning back to the nucleus (abortive transport). The positions of Imp-α/CAS complexes, as determined by smFRET, are indicated by solid purple circles, and the positions of CAS-free Imp-α are identified by blue open circles. The black solid circles identify the last location at which FRET was observed (n = 31 and 20, Upper and Lower, respectively). The orange dashed lines identify the location of Imp-α/CAS complex assembly in the nuclear basket (Left) and Imp-α/CAS complex disassembly in the cytoplasmic filament region (Right). For scale, the green and black lines are −100 and +100 nm from the NE. For all panels, trajectories from multiple NPCs were aligned and overlaid.
Detection of a Transient Cargo/Imp-α/CAS Intermediate During Import Complex Disassembly.
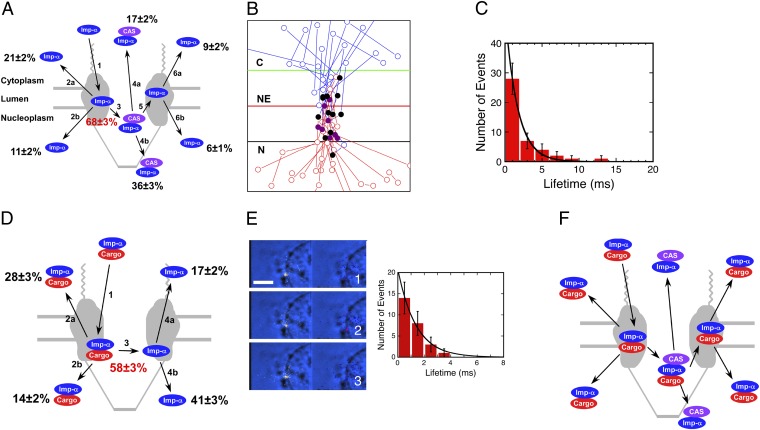
We first used smFRET to follow Imp-α movement from the cytoplasm to the nucleus. Six outcomes were observed when an Imp-α molecule entered an NPC from the cytoplasmic compartment (Fig. 3A), reflecting the spectrum of interactions and dissociation events involved. Only the CAS-free form of Imp-α was observed to interact with NPCs from the cytoplasmic side (Fig. 3A, path 1). smFRET, indicating a binding interaction with CAS, was observed for 68 ± 3% of the interaction events (Fig. 3A, path 3). Imp-α molecules that did not interact with CAS returned to the cytoplasm with ∼2:1 bias over nuclear entry (Fig. 3A, path 2). In contrast, Imp-α/CAS complexes entered the nucleus with ∼2:1 bias over return to the cytoplasm (Fig. 3A, path 4). However, some (15 ± 2%) Imp-α/CAS complexes dissociated at the NPC with a lifetime of 1.7 ± 0.1 ms [Fig. 3 A (path 5), B, and C and Movie S3]. The free Imp-α generated in this way entered either the cytoplasm or the nucleus with an ∼3:2 bias (Fig. 3A, path 6).
Fig. 3.
Quantification of possible outcomes during nuclear import and identification of a transient trimeric cargo/Imp-α/CAS intermediate. (A) The six outcomes observed with Alexa568-Imp-α (blue) and Alexa647-muCAS (purple) during nuclear import. Only the CAS-free form of Imp-α was observed to interact with NPCs from the cytoplasmic side (path 1). Sometimes Imp-α left the NPC without binding to CAS (path 2), and sometimes binding to CAS occurred (path 3). Imp-α/CAS complex formation occurred on the nucleoplasmic side of the NPC (Fig. 2). The Imp-α/CAS complex either escaped from the NPC intact (path 4) or dissociated before Imp-α left the NPC (paths 5 and 6). The Imp-α shown on the right interacted briefly with CAS and thus was experimentally distinguishable from the Imp-α on the left. The three species on the top result from aborted transport, whereas the three species on the bottom are translocated Imp-α molecules. The efficiency of Imp-α/CAS complex formation (path 3) is indicated in red (n = 287). (B) Import trajectories for Alexa568-Imp-α that only transiently exhibited FRET with Alexa647-muCAS. The positions of Imp-α alone are indicated in blue (before complex formation) and red (after complex formation), and Imp-α/CAS complexes, as determined by smFRET, are identified in purple. The black solid circles identify the first location at which FRET was observed (n = 13). For scale, the green and black lines are −100 and +100 nm from the NE. (C) Lifetime histogram (τ = 1.7 ± 0.1 ms) for the Imp-α/CAS complex observed in B (n = 43). (D) The four outcomes observed for Alexa568-Imp-α (blue) and Alexa647-NLS-2xGFP(4C) (red) during nuclear import. The same experiment as in A is shown, except that Alexa647 was on the cargo instead of on CAS. Imp-α always first interacted with NPCs as part of an Imp-α/cargo complex (path 1). This complex sometimes left the NPC intact (path 2) and sometimes dissociated before Imp-α left the NPC (paths 3 and 4). The efficiency of Imp-α/cargo complex disassembly is given in the center in red (n = 288). (E) Movie frames showing the appearance of smFRET between Alexa568-NLS-2xGFP(4C) (0.1 nM) (yellow, Left) and Alexa647-muCAS (250 nM) (red, Right). The smFRET signal is significantly weaker than that observed in Fig. 1, likely because the cargo and CAS molecules do not directly bind to each other. A lifetime histogram yields τ = 1.4 ± 0.1 ms. These data strongly support the formation of a transient trimeric cargo/Imp-α/CAS complex. Numbers denote time (milliseconds). (Scale bar: 5 μm.) (F) Model of the expected species present during cargo import. See text for details. Conditions are the same as in Fig. 1A, except as noted.
The transient nature of a subset of Imp-α/CAS complexes was unexpected. Studies on the yeast analogs of Nup50 and CAS (Nup2 and Cse1, respectively) have shown that both factors independently accelerate the dissociation of Imp-α/cargo complexes in vitro (25, 27, 28, 32). In the context of the NPC, it was proposed that Nup50 first promotes cargo release from Imp-α in the nuclear basket and then CAS promotes Imp-α release from Nup50 (26). We subsequently showed, however, that both Nup50 and CAS are involved in import complex dissociation (30). Because a low nanomolar concentration of exogenous CAS is essential for dissociation of Imp-α/cargo complexes during nuclear import (30), the endogenous CAS concentration remaining in permeabilized cells is likely to be quite low. Surprisingly, the data in Fig. 3A indicate that Imp-α/CAS complexes form 68 ± 3% of the time, which is greater than the fraction (∼55–61%) of Imp-α/cargo complexes that we previously determined dissociated at NPCs (30). The greater fraction of Imp-α/CAS complexes suggested the possibility that the interaction of CAS with Imp-α/cargo complexes did not always immediately lead to cargo dissociation. Because muCAS was not used in the earlier experiments, we reproduced the Imp-α/cargo dissociation data under identical conditions to those in Fig. 3A. We obtained a dissociation efficiency comparable to that obtained earlier (58 ± 3%). Although the transport bias of Imp-α/cargo complexes and free Imp-α differed by a factor of ∼2 (Fig. 3D), compared with our earlier work (30), this probably reflects the difference between wild-type CAS (earlier data) and muCAS (Fig. 3D).
In total, the data in Fig. 3 A and D indicate that ∼10% more of the total Imp-α molecules (68 ± 3% rather than 58 ± 3%; P < 0.005, two-proportion z-test) interact with CAS than predicted from the number of Imp-α/cargo dissociation events. Consequently, these data suggested the existence of a trimeric cargo/Imp-α/CAS complex that disassembles to generate either an Imp-α/CAS complex + cargo or a cargo/Imp-α complex + CAS. We tested this possibility directly by exploring whether smFRET could be observed between cargo (donor dyes) and CAS (acceptor dyes), and indeed this was observed for 8 ± 3% of cargo interactions with the NPC (Fig. 3E and Movie S4). The lifetime of the cargo/CAS FRET interaction was 1.4 ± 0.1 ms (Fig. 3E). These data support the presence of a transient trimeric cargo/Imp-α/CAS complex. Fig. 3F depicts the predicted species and pathways during cargo import, based on this model.
Nup50 Choreography of Imp-α/CAS Complex Assembly.
Three previously characterized Imp-α mutants that impair binding to cargo, Nup50, or CAS (33, 34) were used to test the model postulated in Fig. 3F. The Imp-α D192K mutation inhibits binding to the NLS-2xGFP cargo; the E396R mutation inhibits binding to Nup50; and the R39D mutation inhibits binding of the Imp-β binding (IBB) domain to CAS (see SI Materials and Methods for a more extensive description). These three mutants were evaluated in import experiments with donor dyes on the mutant Imp-α and acceptor dyes on muCAS. The data are summarized in Fig. 4 and Table S1. Despite an ∼10-fold range in the ability of wild-type Imp-α and the various mutants to form Imp-α/CAS complexes, the overall proportion of Imp-α (either free or bound to cargo) that reached the nuclear compartment remained similar (42–53%; Fig. 5).
Fig. 4.
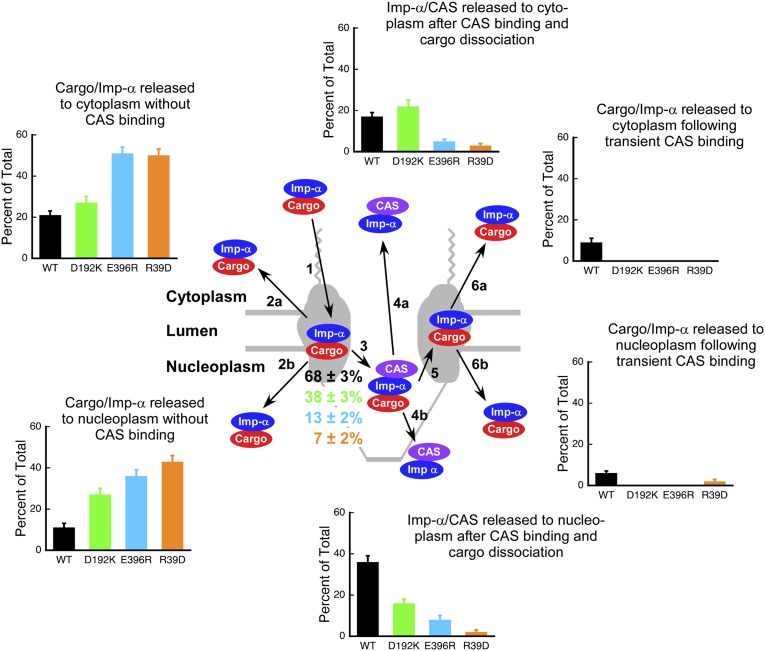
Effect of three Imp-α mutations on nuclear import. The three single-site Imp-α mutations D192K (green), E396R (light blue), and R39D (orange) all reduced the interaction of Imp-α with CAS. Quantified in the bar graphs are the frequency (as percentage of the total) that each of the six outcomes depicted in Fig. 3A was observed for the three mutants and wild-type (WT) Imp-α . Only the CAS-free form of Imp-α was observed to interact with NPCs from the cytoplasmic side (path 1). For wild-type Imp-α, the presence or absence of a cargo molecule was assumed on the basis of the model in Fig. 3F. The cargo is not expected to be present in any of the species depicted for the R39D and D192K mutants (Fig. S2A). Imp-α/CAS complex formation efficiencies (path 3) are indicated as percentages, color coded to the various mutants (n = 207–287; Table S1). Conditions are the same as in Fig. 1A.
Fig. 5.
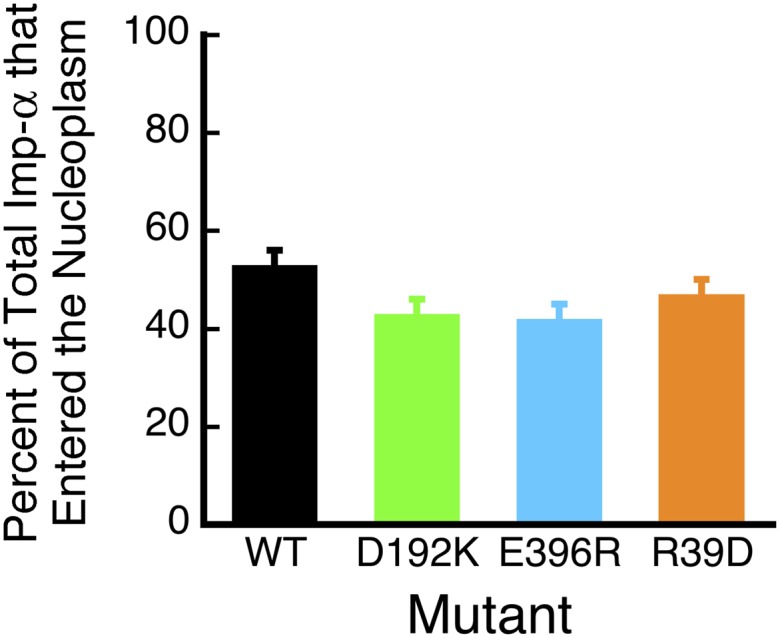
Percentage of the total Imp-α (Imp-α + Imp-α/CAS complexes) released into the nucleoplasm, according to the data in Fig. 4.
The Imp-α mutant data reported here provide strong evidence to support a model in which the dissociation of Imp-α–containing complexes at the nuclear face of NPCs is orchestrated by the nuclear basket nucleoporin Nup50, which choreographs both disassembly of the cargo/Imp-α complex and formation of the Imp-α/CAS complex. Details of the effects of the mutations on the interactions of Imp-α and their implications are summarized in SI Materials and Methods (Fig. S3). Here, we summarize the most important findings for each of the mutants.
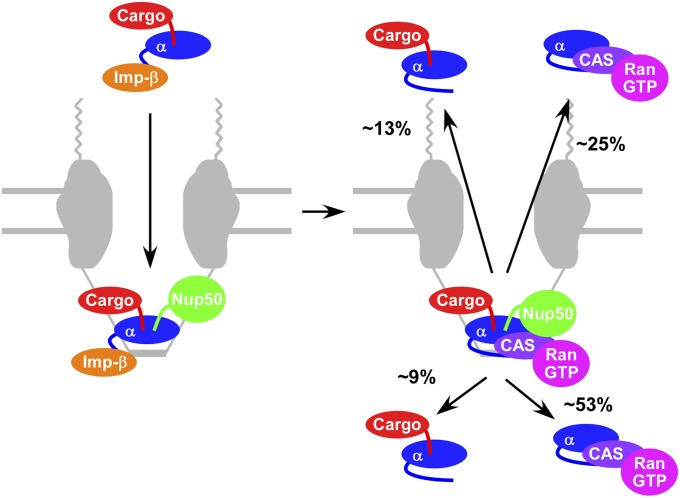
Our results indicate that the likely sequence of events most frequently followed is binding of the cargo/Imp-α/Imp-β complex to Nup50, followed by dissociation of Imp-β and then binding of CAS/RanGTP. This model is supported by the following observations. First, the E396R mutation reduced Imp-α/CAS complex formation by ∼5-fold (from 68 ± 3% to 13 ± 2%; Fig. 4). Because this mutation interferes with the binding of Imp-α to Nup50, Imp-α must usually bind to Nup50 before interacting with CAS. Second, the R39D mutation reduced Imp-α/CAS complex formation by ∼10-fold (from 68 ± 3% to 7 ± 2%; Fig. 4). Because this mutant has a reduced affinity for CAS (Fig. S2) due to the interaction of Imp-α Arg39 with CAS Asp226 (35), these data highlight the importance of the interaction of CAS with the IBB domain. Consequently, they strongly indicate that Imp-β dissociates before Imp-α binds to CAS and that the IBB domain then interacts with CAS. And third, the D192K mutation reduced Imp-α/CAS complex formation by ∼2-fold (from 68 ± 3% to 38 ± 3%; Fig. 4). Because this mutation interferes with the binding of the cargo’s nuclear localization signal (NLS) to Imp-α, these data suggest that the major NLS binding site must be occupied for efficient binding of CAS/RanGTP to Imp-α. The major NLS binding site can be occupied, of course, by the NLS itself, which would suggest that the cargo typically remains bound to Imp-α before it interacts with CAS. The IBB domain can also occupy the major NLS binding site (autoinhibition) (36). However, as the data in Fig. 3E indicate the presence of both cargo and CAS in at least some transient complexes, the NLS–Imp-α interaction remains intact at least some of the time until after Imp-α binds to CAS. As a whole, the Imp-α mutant data suggest that the transient complex we identified consists of the following four or five proteins: cargo, Imp-α, CAS, RanGTP, and Nup50. Fig. 6 summarizes the sequence of events supported by our results. We emphasize that in the experiments reported here, we detected only the cargo, Imp-α, and CAS (in pairwise combinations), and thus our conclusions regarding the involvement of other proteins in various complexes are based on known in vitro affinities, structural constraints, and previous single-molecule experiments, as indicated.
Fig. 6.
Model of the assembly and disassembly of a proposed cargo/Imp-α/CAS/RanGTP/Nup50 complex. Percentages of the four proposed decomposition products are based on Fig. 3A after Imp-α forms a complex with CAS. It is unclear why cargo/Imp-α complexes are preferentially released to the cytoplasm and Imp-α/CAS complexes are preferentially released into the nucleoplasm. See text for details.
Dissociation of Imp-α/CAS Complexes at the Cytoplasmic NPC Face During Export.
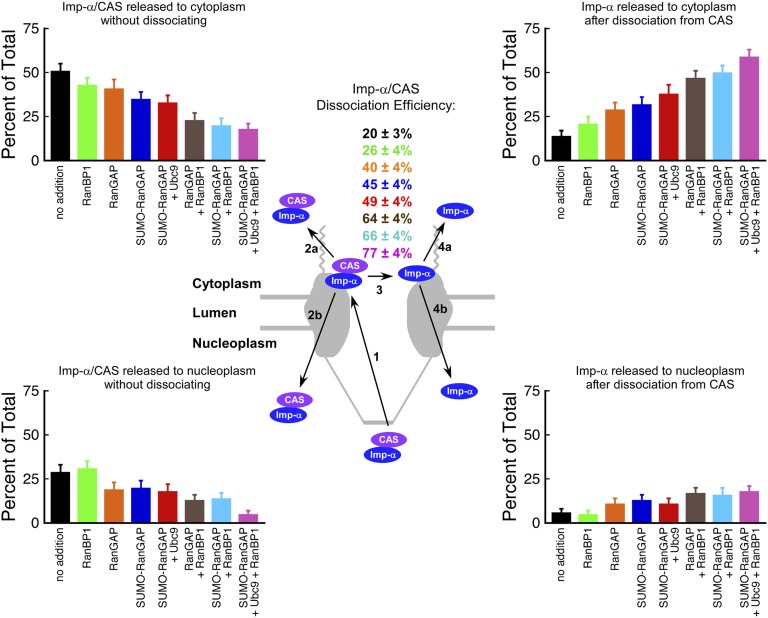
We next used smFRET to follow the dissociation of Imp-α/CAS complexes at the NPC cytoplasmic face following export. Four outcomes were observed when an Imp-α/CAS complex entered an NPC from the nucleoplasm (Fig. 7 and Tables S2 and S3). Here, any endogenous Imp-α (either free or in a complex with fluorescent CAS) was invisible because fluorescence emission was generated only through donor excitation, and thus all detected export complexes contained both fluorescent Imp-α (donor) and fluorescent CAS (acceptor). When an Imp-α/CAS complex entered the NPC from the nucleoplasm, it escaped to the cytoplasm intact, returned to the nucleus, or dissociated, in which case the free Imp-α molecule entered the cytoplasm or the nucleus. Under standard export assay conditions, 20 ± 3% of the Imp-α/CAS complexes dissociated at the NPC (Fig. 7). Dissociation events were centered primarily in the cytoplasmic filament region, ∼100 nm from the center of the nuclear envelope (Fig. 2), consistent with the location where RanGAP is tethered to Nup358 (RanBP2), the major component of metazoan cytoplasmic filaments (37, 38). CAS has a high affinity for Imp-α only when bound to RanGTP (21), as we confirmed in FRET-based bulk binding assays (Fig. S2C). Consequently, although RanGTP was not assayed in our single-molecule experiments, we expect that the Imp-α/CAS complexes included RanGTP. Activation of the Ran GTPase is expected to result in GTP hydrolysis and thereby enhance disassembly of the Imp-α/CAS complex.
Fig. 7.
Effect of Ran GTPase-activating factors on the disassembly of Imp-α/CAS complexes during nuclear export. Four outcomes were observed with Alexa568-Imp-α (blue) and Alexa647-muCAS (purple) during nuclear export. Imp-α/CAS complexes entered NPCs from the nuclear side (path 1). Some Imp-α/CAS complexes left the NPC without dissociating, entering the nucleus or the cytoplasm (path 2). Some Imp-α/CAS complexes dissociated (path 3), and the free Imp-α entered either the nucleus or the cytoplasm (path 4). Dissociation of Imp-α/CAS complexes occurred on the cytoplasmic side of the NPC (Fig. 2). Imp-α/CAS complex dissociation efficiencies (path 3) are indicated in the Center as percentages (n = 112–168; Tables S2 and S3), and the frequency, as a percentage of the total, that each of the four outcomes was observed is indicated by the bar graphs. Black, no addition, i.e., a standard export experiment; green, 3 µM RanBP1; orange, 3 µM RanGAP; blue, 3 µM SUMO1-RanGAP; red, 3 µM SUMO1-RanGAP + 5 µM Ubc9; brown, 3 µM RanGAP + 3 µM RanBP1; light blue, 3 µM SUMO1-RanGAP + 3 µM RanBP1; rose, 3 µM SUMO1-RanGAP + 5 µM Ubc9 + 3 µM RanBP1.
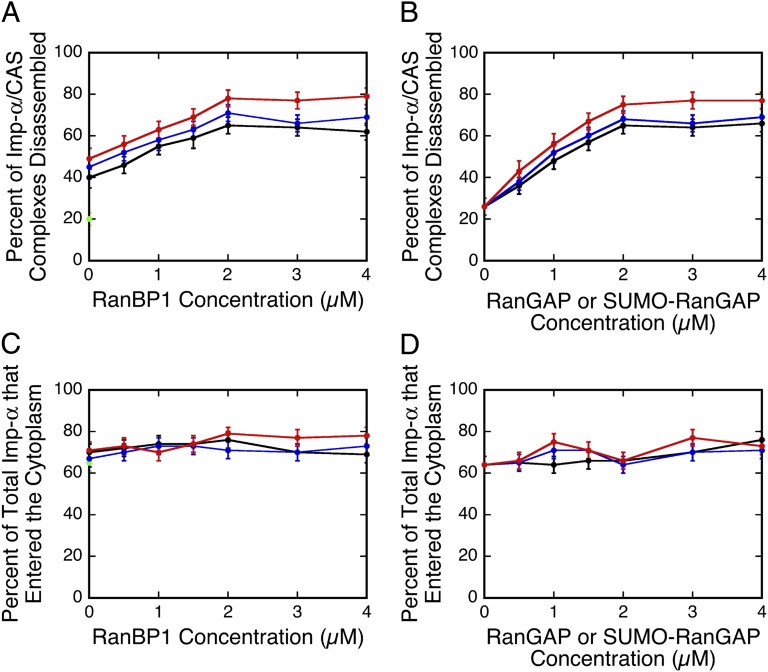
To further test the idea that GTP hydrolysis by Ran leads to Imp-α/CAS complex disassembly, we probed conditions that enhance Ran GTPase activity. Addition of 3 µM RanGAP resulted in an ∼2-fold increase (from 20 ± 3% to 40 ± 4%) in Imp-α/CAS complex dissociation efficiency. We then probed the effect of RanBP1. In vitro, RanBP1 further enhances Ran GTPase activity over that seen with RanGAP alone (39), albeit by binding to Ran without directly contacting RanGAP (40). In the presence of 3 µM RanGAP, RanBP1 increased Imp-α/CAS complex dissociation efficiency by a further ∼1.5-fold, saturating at ∼2 µM RanBP1 (Fig. 8A and Table S2). At 3 µM RanBP1, the Imp-α/CAS complex dissociation efficiency saturates at ∼2 µM RanGAP (Fig. 8B and Table S3). For comparison, the cytoplasmic RanBP1 and RanGAP concentrations in HeLa cells are ∼2 and 0.7 µM, respectively (39, 41). These data are therefore consistent with the hypothesis that RanBP1 promotes Ran GTPase activation by RanGAP.
Fig. 8.
Imp-α/CAS complex disassembly efficiency and total transport of Imp-α. (A) Effect of RanBP1 on Imp-α/CAS disassembly efficiency in the presence of Ran GTPase-activating factors (n = 104–168; Table S2). Green, no addition; black, 3 µM RanGAP; blue, 3 µM SUMO1-RanGAP; red, 3 µM SUMO1-RanGAP + 5 µM Ubc9. (B) Imp-α/CAS disassembly efficiency in the presence of 3 µM RanBP1 and other Ran GTPase-activating factors (n = 116–151; Table S3). Black, RanGAP; blue, SUMO1-RanGAP; red, SUMO1-RanGAP + 5 µM Ubc9. (C and D) Total percentage of Imp-α (Imp-α + Imp-α/CAS complexes) released into the cytoplasm, according to the data in A and B, respectively, using the same coloring scheme. Conditions are the same as in Fig. 1D for A–D, unless otherwise indicated.
RanGAP does not bind directly to Nup358. Rather, the small ubiquitin-like modifier (SUMO) moiety on SUMO-RanGAP interacts with Nup358. Further, Ubc9, the SUMO E2 ligase, enhances this interaction due to contacts with both Nup358 and the RanGAP domain of SUMO-RanGAP (38). SUMOylation of RanGAP had an insignificant effect on Imp-α/CAS complex dissociation efficiency (Fig. 8 A and B). Ubc9 (5 µM) had a small, if any, effect on Imp-α/CAS complex dissociation efficiency in the presence of SUMO-RanGAP (Fig. 8 A and B). The combination of SUMOylation of RanGAP and Ubc9 did significantly increase Imp-α/CAS complex dissociation efficiency in the presence of RanBP1 (Fig. 8A). However, RanBP1 alone was more effective than SUMOylation of RanGAP and Ubc9 in promoting Ran-GAP–dependent Imp-α/CAS complex dissociation (Figs. 7 and 8A). Surprisingly, RanBP1, RanGAP, SUMO-RanGAP, and Ubc9 had essentially no effect on the total fraction of Imp-α that exited into the cytoplasm (Fig. 8 C and D).
Implications of the Effect of Soluble RanGAP on Imp-α/CAS Complex Dissociation at the NPC.
Our data provide insight into the role of RanGAP in the disassembly of the Imp-α/CAS/RanGTP complex after export to the cytoplasm. These data suggest that the soluble, unSUMOylated form of RanGAP can promote Imp-α/CAS complex dissociation at the cytoplasmic face of the NPC during random diffusional encounters when it is not bound to Nup358. An alternative possibility is that exogenous RanGAP becomes SUMOylated in the permeabilized cell assay and hence binds to Nup358. However, yeast RanGAP, which cannot be SUMOylated (42), promotes Imp-α/CAS complex dissociation at NPCs (Fig. S4), supporting the hypothesis that binding of RanGAP to Nup358 is not essential for promoting Imp-α/CAS complex dissociation.
In vitro, dissociation of Ran from CAS and hydrolysis of GTP requires both RanBP1 and RanGAP (19, 21). In our experiments, unSUMOylated RanGAP, which is not thought to bind to NPCs, activated Imp-α/CAS complex disassembly at the NPC following export by ∼2-fold in the absence of added RanBP1, whereas RanBP1 activated dissociation by only ∼1.3-fold in the absence of RanGAP (Fig. 7A). Bischoff and Görlich (19) found that RanBP1 alone strongly promotes dissociation of RanGTP from Imp-α/CAS/RanGTP complexes and proposed that RanGAP-mediated GTP hydrolysis subsequently makes the process irreversible. One explanation for these seeming conflicting data on RanBP1 is that Ran binding domains (RanBPs) on the cytoplasmic filaments (43, 44) can perform the same function as RanBP1, thereby assisting RanGAP in accelerating export complex disassembly at the NPC.
Mathematical Models of Substep Kinetics During Nuclear Import and Export of Imp-α.
Mathematical models were constructed that described the kinetics of the assembly and disassembly of complexes during the nuclear protein import cycle. By synchronizing many single-molecule interaction events to the time point of the initial NPC binding event, pseudofirst-order rate constants were determined for models consisting of a series of first-order reactions by globally fitting the fraction of each species present as a function of time. In the following analyses, we consider only those molecules that both completed transport and underwent assembly/disassembly during transport. We first discuss the kinetics occurring at the termination of nuclear protein import when the cargo dissociates from Imp-α and CAS binds to Imp-α. We then discuss the kinetics of CAS-mediated export of Imp-α and its release into the cytoplasm.
Substep Kinetics During Imp-α–Mediated Nuclear Import.
Two smFRET datasets were analyzed for the import process, each of which assayed for a different pairwise interaction. The three species identified when monitoring Imp-α/CAS complex formation are (A) NPC-bound Imp-α, (B) NPC-bound Imp-α/CAS complex, and (C) the Imp-α/CAS complex free in the nucleoplasm. Data were fitted to an A → B → C model, yielding τ1 (A → B) ∼ 4.2 ms and τ2 (B → C) ∼ 1.3 ms (Fig. S5A). The three identified species when monitoring dissociation of the Imp-α/cargo complex are (A) NPC-bound Imp-α/cargo complex, (B) NPC-bound cargo-free Imp-α, and (C) cargo-free Imp-α in the nucleoplasm. For this reaction sequence, data were also fitted to an A → B → C model, yielding τ1 (A → B) ∼ 4.4 ms and τ2 (B → C) ∼ 1.0 ms (Fig. S5B).
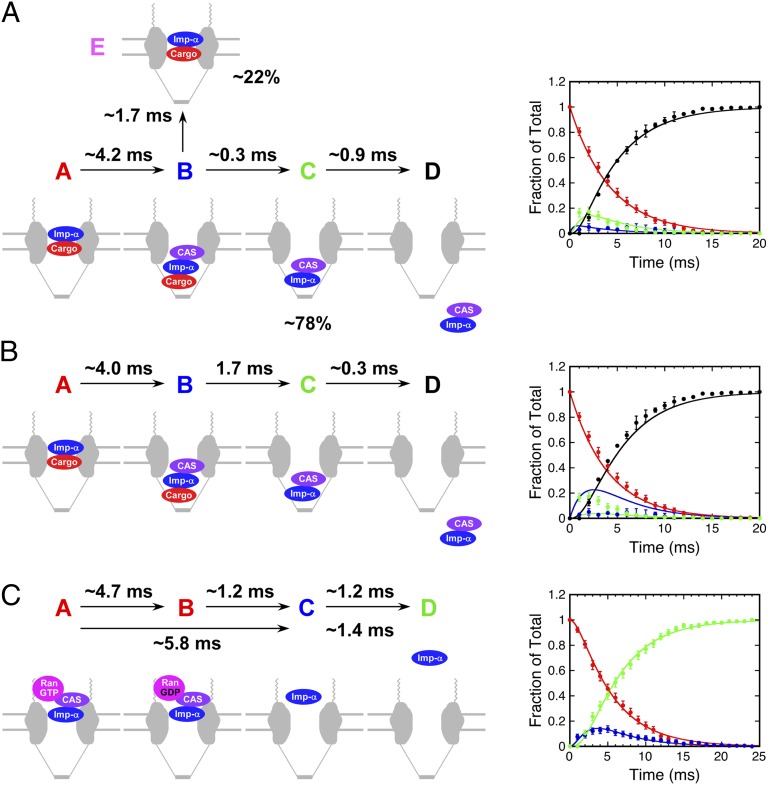
The identification of a trimeric cargo/Imp-α/CAS complex during import indicates, however, that the two-step first-order models fitted to the data, as discussed in the preceding paragraph, are overly simplistic. Because the Imp-α/CAS FRET signal is expected to be similar for cargo/Imp-α/CAS and Imp-α/CAS complexes, the trimeric intermediate is indistinguishable from the dimeric species in an individual dataset. We addressed this uncertainty by combining the kinetic data in Fig. S5 A and B, assuming the following four species: (A) NPC-bound cargo/Imp-α complex, (B) NPC-bound cargo/Imp-α/CAS complex, (C) NPC-bound Imp-α/CAS complex, and (D) Imp-α/CAS complex free in the nucleoplasm. The data in Fig. S5A represent species A + B, C, and D, whereas the data in Fig. S5B represents species A, B + C, and D. The populations of species A, C, and D were obtained directly from the datasets (the D data were averaged). The population of species B was calculated twice [B = (A + B) – A and B = (B + C) – C] and averaged. Global fitting revealed a very short lifetime (∼0.3 ms) for the trimeric cargo/Imp-α/CAS complex (Fig. 9A).
Fig. 9.
Kinetics of complex assembly and disassembly. All reactions were modeled as three-step first-order processes, A → B → C → D, and data were fitted using Berkeley Madonna. (A) Assembly of Imp-α/CAS complexes during nuclear import. Data were pooled/calculated from Fig. S5 A and B. The 1.7 ± 0.1-ms lifetime observed for some Imp-α/CAS complexes (Fig. 3C) can be explained as an off-pathway decomposition reaction of cargo/Imp-α/CAS complexes. Percentages reflect the decomposition from the total cargo/Imp-α/CAS pool, according to the data and model in Fig. 3 A and F. (B) Same reaction and model as in A, assuming that the time constant for the second step is 1.7 ms. See text for details. (C) Disassembly of Imp-α/CAS complexes during nuclear export. The upper time constants are from fitting to a three-step model (shown in the graph) and the lower time constants are from fitting to the two-step model (Fig. S5C). Data are from Fig. S5C. See text for details.
Implications of the Kinetic Analysis of Imp-α/CAS Complex Formation During Nuclear Import.
The lifetime of the trimeric cargo/Imp-α/CAS complex obtained from the kinetic analysis (∼0.3 ms; Fig. 9A) was significantly shorter than the 1.7 ± 0.1-ms lifetime obtained from the Imp-α/CAS complex data (Fig. 3C). This difference likely arises because the 1.7-ms lifetime results only for cargo/Imp-α/CAS complexes that yield cargo/Imp-α complexes and free CAS, whereas the 0.3-ms lifetime reflects the decomposition of cargo/Imp-α/CAS complexes to yield free cargo and Imp-α/CAS complexes (Fig. 9A). The first decomposition reaction is not reflected in the kinetic dataset, which includes only species involved in complete transport. The short ∼0.3-ms lifetime is supported by the low frequency with which the cargo–CAS FRET interaction was detected directly (8 ± 3%); i.e., most such FRET signals were not observed due to decomposition before detection. The 0.3- and 1.7-ms decay constants predict that ∼85% of molecules decay by the faster pathway ([C]/[E] = 1.7 ms/0.3 ms; Fig. 9A), consistent with the 78% partitioning that was observed (Fig. 3A). The predicted lifetime of the cargo/Imp-α/CAS intermediate considering the two predicted decay pathways is ∼0.3 ms (τ = (1/τ1 + 1/τ2)−1). This value is significantly shorter than the observed 1.4-ms lifetime of the FRET interaction between CAS and cargo (Fig. 3E). However, the shorter lifetime species is likely highly underrepresented in the observed cargo/CAS complex lifetime dataset due to the 1-ms time resolution of the measurements and the weak FRET signal. The robustness of the global kinetic fit is demonstrated by fixing the time constant for cargo/Imp-α/CAS complex decomposition at 1.7 ms, i.e., assuming that cargo and CAS dissociate at similar rates. With this assumption, the global fitting is significantly worse (Fig. 9B). We therefore conclude that the 1.7-ms lifetime obtained in Fig. 3C does not reflect the dominant decay pathway of the cargo/Imp-α/CAS intermediate.
Substep Kinetics During the Nuclear Export of Imp-α Bound to CAS.
From smFRET Imp-α export data, we identified three distinguishable species: (A) NPC-bound Imp-α/CAS complex, (B) NPC-bound Imp-α, and (C) Imp-α in the cytoplasm. Data were fitted to an A → B → C model, yielding τ1 (A → B) ∼ 5.8 ms and τ2 (B → C) ∼ 1.4 ms (Fig. S5C). The fit to the data for the A and B intermediates was reasonable, but, because more is known about this process, we tested a more complex model that includes a GTP hydrolysis step. Thus, we assumed a four-species model, A → B → C → D, with (A) NPC-bound Imp-α/CAS/RanGTP complex, (B) NPC-bound Imp-α/CAS/RanGDP (or Imp-α/CAS) complex, (C) NPC-bound Imp-α, and (D) Imp-α in the cytoplasm. For this model, species A and B are indistinguishable within the dataset. This is accommodated within the fitting protocol by assuming that the detected Imp-α/CAS complex is the sum of species A and B. For this model, τ1 (A → B) ∼ 4.7 ms, τ2 (B → C) ∼ 1.2 ms, and τ3 (C → D) ∼ 1.2 ms (Fig. 9C).
The time constant for the last step, escape of Imp-α from the NPC, is approximately the same for both models discussed in the previous paragraph. The fit obtained from the four-step model suggests that activation of the Ran GTPase is the slower step (A → B) and the Imp-α/CAS complex dissociates reasonably quickly following GTP hydrolysis (B → C). This is not unexpected, because the Ran GTPase activation step includes diffusion of the Imp-α/CAS complex through and within the FG network and finding and binding to RanGAP (pseudofirst order), whereas the disassembly step is a unimolecular decomposition (true first order).
Total Imp-α Released into the trans-Compartment Was Surprisingly Constant for both Import and Export.
The proportion of the total Imp-α released into the nucleus and the cytoplasm was not influenced substantially by Imp-α mutations and Ran GTPase activation (Figs. 4, 5, 7, and 8). These observations are consistent with our earlier observation that the proportion of Imp-α (free or bound to cargo) released into the nucleus was a surprisingly consistent ∼50% over a range of CAS concentrations in which the cargo/Imp-α dissociation efficiency ranged from 0 to ∼60% (30). Thus, the proportion of Imp-α (free or bound to cargo or CAS) released into the nucleus must be determined primarily by RanGTP dissociating Imp-β from the import complex. After dissociation of Imp-α from Imp-β, the interaction of Imp-α with Nup50 and CAS promotes dissociation of the cargo from Imp-α, resulting in productive nuclear import, as well as allowing Imp-α to be exported to participate in another import cycle. A similar effect was observed for Imp-α/CAS complexes undergoing export. Export complex disassembly efficiency varied by approximately fourfold (Fig. 7) and yet the proportion of Imp-α (free or complexed with CAS) released into the cytoplasm (64–76%) was largely unchanged (Fig. 8 C and D). Because a three-step model, rather than a two-step model, is needed to adequately describe the Imp-α/CAS complex dissociation reaction, a transient intermediate exists (τ ∼ 1 ms), which is probably an Imp-α/CAS/RanGDP (or Imp-α/CAS) complex. The ∼2:1 biased release of Imp-α into the cytoplasm independent of soluble Ran GTPase-activating proteins (Fig. 8 C and D) indicates that the export complex must be altered in some way before encountering these proteins, likely resulting from the binding of the Imp-α/CAS complex to RanBP2 (predicting at least a four-step model). Therefore, Ran GTPase activation by soluble proteins facilitates Imp-α/CAS complex dissociation so that Imp-α may begin another round of import but does not drive release of Imp-α from the NPC.
Implications for the Nuclear Protein Import Cycle.
Our smFRET data have uncovered features of key steps in the nuclear protein import cycle and extend the general picture developed from previous studies in which the cytoplasmic and nucleoplasmic faces of NPCs contain factors that promote transport complex disassembly and assembly concomitant with transport through the NPC (reviewed by refs. 15 and 17). Because our approach monitors interaction sequences in the context of intact NPCs, it ensures the spatiotemporal preservation of motifs and binding sites critical for activity in vivo. Notably, all of the association/dissociation events and transport processes that we observed were sampled only by a fraction of the molecules involved in the nuclear protein import cycle. Three general features of NPC transport reactions are highlighted by our single-molecule approach: First, key interactions between the components are reversible; second, multiple outcomes are often possible; and third, assembly and disassembly dynamics occur on the timescale of the transport reactions (milliseconds). These properties indicate that the transport reactions operate near thermodynamic and kinetic equilibrium, which, in turn, implies that small perturbations can be significantly disruptive. Crucially, our real-time studies in intact NPCs show that transport complex assembly and disassembly occur orders of magnitude faster than they do in vitro (milliseconds vs. tens of seconds or more), resulting in significantly different results and interpretations.
Our results provide direct experimental support for the presence of distinct environments at the nuclear and cytoplasmic faces of functioning NPCs, as evidenced by distinct spatial regions that orchestrate the assembly and disassembly of transport complexes, and give insight into the roles of Nup50 and RanGAP. The transient cargo/Imp-α/CAS complex identified by smFRET has not previously been considered when analyzing nuclear import, and this observation imposes kinetic and structural constraints on the binding interactions. smFRET also demonstrated that the unSUMOylated form of RanGAP, which is not thought to bind directly to NPCs, unexpectedly increases the dissociation efficiency of Imp-α/CAS export complexes at the NPC. Although the intricate series of interactions identified here illustrates the complexity of nuclear transport, analysis of the kinetics of the process has enabled the development of quantitative models that describe the system well and that have powerful predictive capability.
Materials and Methods
Plasmids and Proteins.
NLS-2xGFP(4C), Imp-β, Imp-α, Ran, nuclear transport factor 2 (NTF2), RanBP1, RanGAP, and CAS were expressed and purified as in our earlier studies (30, 31, 45, 46). Human Imp-α mutants D192K and E396R were generated on the basis of previous characterizations (34) and the R39D Imp-α mutant was constructed on the basis of the corresponding (R44D) mutation in yeast karyopherin α (35). All Imp-α and CAS mutations were made by the QuikChange protocol (Stratagene) and confirmed by DNA sequencing. Uncleaved GST-muCAS was used for binding experiments only (Fig. S2). The solvent-accessible cysteines on NLS-2xGFP(4C), Imp-α, CAS, and muCAS were reacted with a 20-fold molar excess of maleimide dye (Invitrogen) for 2 h. Labeling ratios of ∼3.5, ∼4, ∼3.4, and ∼5.3 dye molecules per protein molecule, respectively, were obtained from single-molecule photobleaching histograms (46). SUMO-RanGAP was generated via in vivo SUMOylation (47) and purified by the same approach as was used for RanGAP (48). Ubc9 was purified as described in ref. 49. CAS and muCAS concentrations were determined by densitometry of Coomassie-stained SDS/PAGE gels, using BSA as a standard. Other protein concentrations were determined by the bicinchoninic acid method (Pierce), using BSA as a standard.
Single-Molecule Approaches.
Our general single-molecule approaches have been described (30, 31, 45, 46). The methodology to monitor oligomerization state by smFRET during nucleocytoplasmic transport in permeabilized HeLa cells (30) was used here with minor modifications. DTT was included in the transport buffer [20 mM Hepes, pH 7.3, 1.5% (wt/vol) polyvinylpyrrolidone (360 kDa) 110 mM KOAc, 5 mM NaOAc, 2 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT] and all experiments were done in the absence of glycerol (used previously to slow diffusion). The Alexa568 dye was excited with either a 568-nm or a 561-nm laser line. For Imp-α labeled with four Alexa568 dye molecules, the time for the second of four (or first of three) dyes to photobleach was ≥140 ms, as observed earlier (30). To simplify single-molecule tracking, short region of interest movies were extracted from raw data movies. Although the CAS protein was present at a relatively high concentration, it was not detectable unless it formed a FRET complex with Imp-α (the protein with the donor dyes). This can be clearly seen from the low background in the acceptor channel. It is certainly possibly that more than one CAS protein was present in a single NPC. However, we never observed the gain of FRET, the loss of FRET, and the regain of FRET, which could be indicative of a second binding event. It is possible that such a process occurred within our time resolution, but we consider this unlikely.
Imp-α Export Assay.
Nuclei of permeabilized cells were loaded with Imp-α for 2 min by addition of 0.1 nM Alexa568-Imp-α, 0.5 μM Imp-β, 2 μM Ran, 1 μM NTF2, and 1 mM GTP. Then, 250 nM Alexa647-muCAS with 2 μM Ran, 1 μM NTF2, and 1 mM GTP were added to initiate export. For single-molecule assays, images were acquired at 1,000 frames per second during the 1- to 2-min time window after CAS addition.
Binding Equation.
Binding affinities were determined from titration experiments using FRET signals, assuming
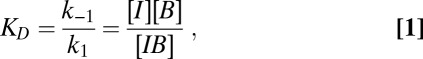 |
where I is Imp-α (with acceptor dyes) and B is a binding partner (with donor dyes). If B0 = [B] + [BI] is the total concentration of the binding partner added, I0 is the added Imp-α concentration, and γ is the fraction of free unbound B, then the total fluorescence of the donor dyes is given by Ftot = γFB + (1 − γ)FIB, where FB and FIB are the fluorescence of unbound and bound B, respectively. By definition, γ = [B]/B0 = (B0 − [BI])/B0. [BI] is obtained as described earlier (50), and substitution yields
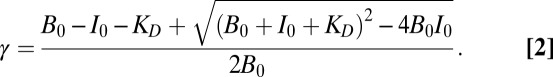 |
For normalized data (FB = 1), there are two fitting parameters, FIB and KD.
Errors.
The position of (dis)assembly events (Fig. 2) and ensemble FRET and KD values (Fig. S2) are reported as mean ± SEM. For complex (dis)assembly events or outcomes reported as a percentage of the total, the errors reported are the SEs for the proportions in a binomial distribution, SE = (p(1 − p)/N)1/2, where p is the measured frequency of the outcome of interest and N is the total number of measurements. The error bars in Fig. 3 C and E are N1/2.
Supplementary Material
Acknowledgments
We thank H. Saitoh for the bacterial SUMOylation system and F. Melchior for the Ubc9 expression plasmid. We especially thank K. Watanabe for pointing out the absence of DTT in the transport buffer used in previous work. This work was supported by the National Institutes of Health (GM065534 and GM084062), the Department of Defense (N00014-02-1-0710), the Welch Foundation (BE-1541), Medical Research Council Grant U105178939, and a Wellcome Trust Programme grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.R.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220610110/-/DCSupplemental.
References
- 1.Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20(6):1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurt E, et al. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275(12):8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- 3.Richardson WD, Mills AD, Dilworth SM, Laskey RA, Dingwall C. Nuclear protein migration involves two steps: Rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52(5):655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- 4.Fahrenkrog B, Aebi U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4(10):757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 5.Stoffler D, et al. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J Mol Biol. 2003;328(1):119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 6.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276(20):16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 7.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158(5):915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100(5):2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rout MP, et al. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148(4):635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6(3):197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 11.Tran EJ, Wente SR. Dynamic nuclear pore complexes: Life on the edge. Cell. 2006;125(6):1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28(17):2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naim B, Zbaida D, Dagan S, Kapon R, Reich Z. Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 2009;28(18):2697–2705. doi: 10.1038/emboj.2009.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colwell LJ, Brenner MP, Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLoS Comput Biol. 2010;6(4):e1000747. doi: 10.1371/journal.pcbi.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8(3):195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 16.Jamali T, Jamali Y, Mehrbod M, Mofrad MRK. Nuclear pore complex: Biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int Rev Cell Mol Biol. 2011;287:233–286. doi: 10.1016/B978-0-12-386043-9.00006-2. [DOI] [PubMed] [Google Scholar]
- 17.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 18.Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15(20):5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 19.Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419(2–3):249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 20.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91(7):2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90(6):1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 22.Tu L-C, Musser SM. Single molecule studies of nucleocytoplasmic transport. Biochim Biophys Acta. 2010;1813:1607–1618. doi: 10.1016/j.bbamcr.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25(3):327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Kelly SM, Corbett AH. Messenger RNA export from the nucleus: A series of molecular wardrobe changes. Traffic. 2009;10(9):1199–1208. doi: 10.1111/j.1600-0854.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilchrist D, Rexach M. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin α in the nucleoplasm. J Biol Chem. 2003;278(51):51937–51949. doi: 10.1074/jbc.M307371200. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24(21):3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol Cell Biol. 2000;20(22):8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 2003;22(20):5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu SM, Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-β homologue, Kap95p. J Mol Biol. 2005;349(3):515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Sun C, Yang W, Tu L-C, Musser SM. Single-molecule measurements of importin α/cargo complex dissociation at the nuclear pore. Proc Natl Acad Sci USA. 2008;105(25):8613–8618. doi: 10.1073/pnas.0710867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Musser SM. Visualizing single molecules transiting through nuclear pore complexes with narrow-field epifluorescence microscopy. Methods. 2006;39:3316–3328. doi: 10.1016/j.ymeth.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilchrist D, Mykytka B, Rexach M. Accelerating the rate of disassembly of karyopherin.cargo complexes. J Biol Chem. 2002;277(20):18161–18172. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- 33.Gruss OJ, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 2001;104(1):83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 34.Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J Biol Chem. 2010;285(23):17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432(7019):872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 36.Catimel B, et al. Biophysical characterization of interactions involving importin-α during nuclear import. J Biol Chem. 2001;276(36):34189–34198. doi: 10.1074/jbc.M103531200. [DOI] [PubMed] [Google Scholar]
- 37.Walther TC, et al. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158(1):63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14(4):705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seewald MJ, Körner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415(6872):662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 41.Görlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22(5):1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA. 2001;98(26):15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokoyama N, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376(6536):184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270(23):14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 45.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101(35):12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin β concentration. J Cell Biol. 2006;174(7):951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchimura Y, Nakamura M, Sugasawa K, Nakao M, Saitoh H. Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal Biochem. 2004;331(1):204–206. doi: 10.1016/j.ab.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 48.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16(6):1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner A, Moutty MC, Möller U, Melchior F. Performing in vitro sumoylation reactions using recombinant enzymes. Methods Mol Biol. 2009;497:187–199. doi: 10.1007/978-1-59745-566-4_12. [DOI] [PubMed] [Google Scholar]
- 50.Whitaker N, Bageshwar UK, Musser SM. Kinetics of precursor interactions with the bacterial Tat translocase detected by real-time FRET. J Biol Chem. 2012;287(14):11252–11260. doi: 10.1074/jbc.M111.324525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.