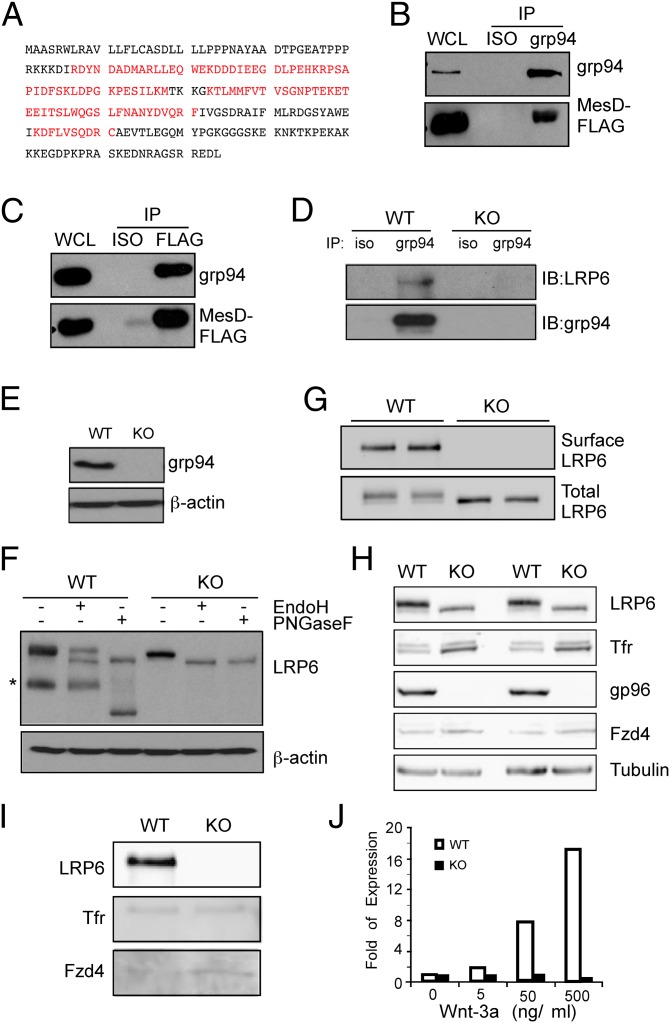
Fig. 1.
grp94 interacts with MesD and is a critical molecular chaperone for LRP6. (A) Identification of MesD as a grp94-interacting protein. MS/MS analysis of grp94-interacting proteins identified MesD. The sequence in red reflects regions authenticated by MS. (B) Immunoblot of grp94 and MesD-FLAG following immunoprecipitation of MesD-FLAG from HEK293 cell lysates. (C) Immunoblot of grp94 and MesD-FLAG following immunoprecipitation of grp94 from HEK293 cell lysates. Immunoprecipitation with isotype control antibody (ISO) is also shown. Expression of grp94 and MesD in whole cell lysate (WCL) is indicated. Data are representative of two independent experiments. (D) Immunoblot of LRP6 and grp94 following immunoprecipitation of grp94 from lysates of WT or grp94 KO MEFs. (E) Immunoblot analysis of grp94 from WT and grp94 KO MEF lysates. (F) Expression of endogenous LRP6 and its sensitivity to N-glycase Endo H and peptide N glycosidase F (PNGaseF) in WT and grp94 KO MEFs. Asterisk indicates cleaved LRP6. β-Actin served as a loading control. (G) WT or grp94 KO MEFs (duplicates) were subjected to surface biotinylation and pull-down with avidin beads, followed by immunoblot for cell surface LRP6. Total LRP6 was also blotted as a control. (H) Immunoblot of various proteins from the total lysates of WT and grp94 MEFs. After incubation of WT and grp94 mutant cells with EZ-Link Sulfo-NHS-SS-Biotin, lysates from cells were examined by immunoblot analysis for LRP6, transferrin receptor (Tfr), grp94, Fzd4, and tubulin. (I) Cell surface biotinylation of WT and grp94 KO MEFs. Lysates from cells treating with EZ-Link Sulfo-NHS-SS-Biotin were immunoprecipitated with NeutrAvidin beads and harvested. The pull-down–biotinylated membrane surface proteins were analyzed by immunoblotting. (J) Expression of Wnt target gene, Axin2, in response to Wnt stimulation. WT and grp94 KO MEFs were stimulated with Wnt-3a for 24 h, and fold expression of the Axin2 relative to unstimulated cells was measured by quantitative PCR. Data are representative of more than two independent experiments.