Abstract
Th17 cells are highly proinflammatory cells critical for clearing extracellular pathogens and for induction of multiple autoimmune diseases1. IL-23 plays a critical role in stabilizing and reinforcing the Th17 phenotype by increasing expression of IL-23 receptor (IL-23R) and endowing Th17 cells with pathogenic effector functions2, 3. However, the precise molecular mechanism by which IL-23 sustains the Th17 response and induces pathogenic effector functions has not been elucidated. Here, we used transcriptional profiling of developing Th17 cells to construct a model of their signaling network and nominate major nodes that regulate Th17 development. We identified serum glucocorticoid kinase-1 (SGK1), a serine-threonine kinase4, as an essential node downstream of IL-23 signaling. SGK1 is critical for regulating IL-23R expression and stabilizing the Th17 cell phenotype by deactivation of Foxo1, a direct repressor of IL-23R expression. SGK1 has been shown to govern Na+ transport and salt (NaCl) homeostasis in other cells5, 6, 7, 8. We here show that a modest increase in salt concentration induces SGK1 expression, promotes IL-23R expression and enhances Th17 cell differentiation in vitro and in vivo, accelerating the development of autoimmunity. Loss of SGK1 abrogated Na+-mediated Th17 differentiation in an IL-23-dependent manner. These data demonstrate that SGK1 plays a critical role in the induction of pathogenic Th17 cells and provides a molecular insight into a mechanism by which an environmental factor such as a high salt diet triggers Th17 development and promotes tissue inflammation.
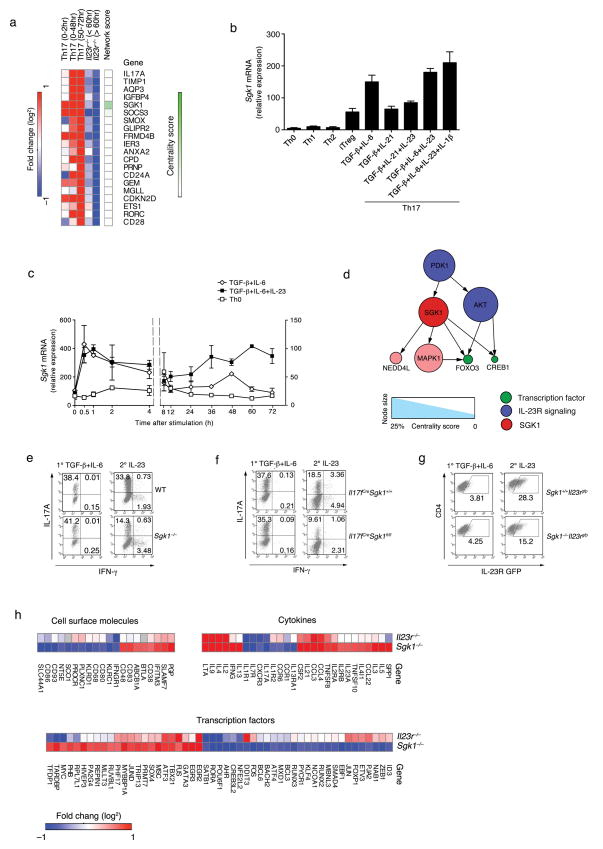
To determine the molecular mechanisms by which naïve T cells develop into effector Th17 cells, we measured genome-wide mRNA expression profiles using microarrays along 18 time points over 72 hours following the in vitro exposure of naive T cells to Th17 polarizing conditions (TGF-β1 with IL-6). To examine the role of IL-23 in Th17 development, we added IL-23 at the late time points (48 – 72 h) and monitored the transcriptional response in both wild-type (WT) and Il23r−/− cells. We ranked the genes according to their extent of induction in cells treated with TGF-β1 and IL-6 (relative to nonpolarized activated T cells) and repression in Il23r−/− cells (relative to WT cells) (Methods; Fig. 1a and Supplementary Table 1). SGK1 was one of the top ranking genes, whose transcriptional regulation is strongly associated with both IL-23R signaling and Th17 cell differentiation (Fig. 1a). qPCR analysis showed that SGK1 is induced at low levels by TGF-β1 (iTreg), and not induced in other T cell subsets (Th0, Th1, Th2). As expected, it is most highly expressed under Th17 differentiation conditions (Fig. 1b). SGK1 expression is strongly induced during the first two hours following stimulation of naïve T cells under Th17-polarizing conditions. This is followed by a sharp decline by 10 hours to a steady expression level that is still substantially higher than in the control population (Fig. 1c and Supplementary Fig. 1a). Furthermore, SGK1 expression is specifically induced and maintained by exposure to IL-23 (Fig. 1c and Supplementary Fig. 1b). While Il23r−/− T cells initially produce SGK1 mRNA, they cannot sustain this expression (Supplementary Fig. 1b, c). Finally, the kinase activity of SGK1 is also significantly higher in Th17 cells than in other T cell subsets (Supplementary Fig. 1d), and restimulation of differentiated Th17 cells with IL-23 further elevates SGK1 kinase activity (Supplementary Fig. 1e). Thus, IL-23 signaling is critical for maintaining SGK1 expression during Th17 cell differentiation.
Figure 1.
Network analysis9 of the transcriptional changes in Il23r−/− T cells singled out SGK1 as a potential nodal point downstream of IL-23R signaling. Based on a curated database of protein-protein interactions (PPI), we constructed a network model that connects known proteins of the IL-23R signaling pathway (Methods) to the transcription factors whose function is dysregulated in Il23r−/− cells (Methods; Fig. 1d and Supplementary Fig. 1f). We ranked the network’s nodes based on a centrality measure, defined as the fraction of IL-23R-affected transcription factors downstream of that node in the network (Methods; Supplementary Table 1). SGK1 was the highest-ranking node (Supplementary Fig. 1g), suggesting that it acts both as a transcriptional target of IL-23R signaling and as a kinase that may mediate the transcriptional effects of the pathway.
Using Sgk1−/− mice, we studied the impact of loss of SGK1 on Th17 differentiation in vitro. We observed no abnormality of SGK1-deficient T cells during primary differentiation into Th17 cells (Fig. 1e). However, Sgk1−/− Th17 cells restimulated with IL-23 showed impaired IL-17 production (Fig. 1e and Supplementary Fig. 2b). Memory Sgk1−/− T cells also showed a defect in IL-17 production upon IL-23 stimulation, but not under stimulation with TGF-β1 and IL-6 (Supplementary Fig. 2a). To study the function of SGK1 specifically in IL-17-producing CD4+ T cells, we generated Il17fCreSgk1fl/fl mice. Il17fCreSgk1fl/fl T cells also showed no defect in primary Th17 differentiation, but displayed reduced IL-17 production upon restimulation with IL-23 (Fig. 1f and Supplementary Fig. 2c). IL-23R expression was also significantly reduced in Sgk1−/− T cells (Fig. 1g). Thus, loss of SGK1 does not affect primary Th17 differentiation, but profoundly affects their stability and IL-23R expression. One possible explanation for the dispensability of SGK1 during primary Th17 differentiation is redundancy with other kinases, such as its homolog AKT10. However, SGK1 seems to be indispensable for IL-23R-dependent stability and maintenance of Th17 cells.
Microarray analysis of Sgk1−/− vs. WT Th17 cells restimulated with IL-23 showed a significant overlap in differentially expressed genes with the Il23r−/− vs. WT IL-23-restimulated Th17 cell profiles, further supporting the functional relatedness of the SGK1 and IL-23R pathways (Fisher exact test, p<10−3) (Fig. 1h and Supplementary Fig. 2d). Consistently, genes downregulated in Sgk1−/− cells are significantly enriched (Fisher exact test, p<10−6) for genes that are upregulated in WT Th17 cells compared to other T cell subsets11 (Methods and Supplementary Fig. 2e), Selected genes were confirmed by qPCR analysis (Supplementary Fig. 2f). Genes from several other pathways are also enriched (over- or underexpressed) (Supplementary Table 2), including cell cycle and proliferation, which may be related to the known role of SGK1 as a regulator of proliferation and apoptosis7, 8, 10. Although our analysis strongly associates SGK1 with the Th17 program, genes important for development and function of other T cell subsets, such as Ifng, Tbx21 or Gata3 were also dysregulated in Sgk1−/− cells, suggesting possible additional effects of this kinase in other T cell subsets.
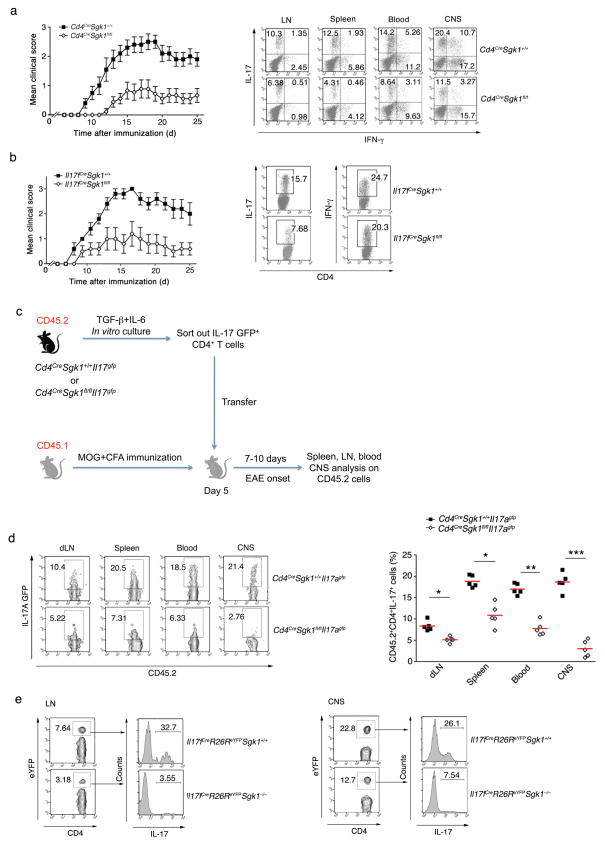
To determine the role of SGK1 in vivo, we immunized Cd4CreSgk1fl/fl mice with MOG35–55 to induce experimental autoimmune encephalomyelitis (EAE). SGK1-deficient mice exhibited significantly reduced EAE incidence and severity. IL-17 production from infiltrated CD4+ T cells in different organs of SGK1-deficient mice was also reduced, whereas IFN-γ levels were unaffected (Fig. 2a and Supplementary Fig. 3a). When we restimulated the isolated T cells from immunized mice with IL-23 in the presence of MOG35–55, the SGK1-deficient T cells also exhibited impaired IL-17 but normal IFN-γ production (Supplementary Fig. 3b, c). Next, using Il23rgfp reporter mice, we observed reduced IL-23R (GFP) expression on infiltrating CD4+ T cells in different organs of SGK1-deficient mice undergoing EAE (Supplementary Fig. 4a). Consistent with Cd4CreSgk1fl/fl mice, reduced Th17 differentiation and disease severity were also observed in Il17fCreSgk1fl/fl mice during EAE (Fig. 2b). In addition, to exclude any effects of SGK1-deficient bystander cells, we transferred purified Il17fCreSgk1fl/fl CD4+ T cells into Rag2−/− mice and induced EAE. Mice that received SGK1-deficient T cells developed attenuated disease as compared to mice that received WT T cells (Supplementary Fig. 4b).
Figure 2.
To determine why we observed fewer Th17 cells in SGK1-deficient mice, we transferred purified GFP+ cells from differentiated Cd4CreSgk1fl/flIl17agfp or control Th17 cells to congenic Ly5.1 mice and traced the IL-17 GFP+ cells in different organs following immunization with MOG35–55 (Fig. 2c). Starting with the same number of CD4+IL-17+ T cells, we found that 7 and 12 days after transfer, SGK1-deficient Th17 cells failed to maintain IL-17 production, especially in the central nervous system (CNS) (Fig. 2d and Supplementary Fig. 4c). Next, we crossed Il17fCreR26ReYFP mice onto the SGK1-deficient background, and analyzed the expression of IL-17 in T cells that had turned on the Il17f gene as determined by eYFP-expression. We induced EAE in these mice and analyzed the frequency of eYFP+ cells producing IL-17 in infiltrating CD4+ T cells in the lymph nodes (LN) and CNS. The Sgk1−/− reporter mice exhibited a smaller proportion of CD4+eYFP+ T cells in both organs. Furthermore, there was a dramatic loss of IL-17 expression by eYFP+ T cells in the SGK1-deficient mice, indicating that Th17 cells could not stably retain IL-17 production during EAE (Fig. 2e and Supplementary Fig. 4d).
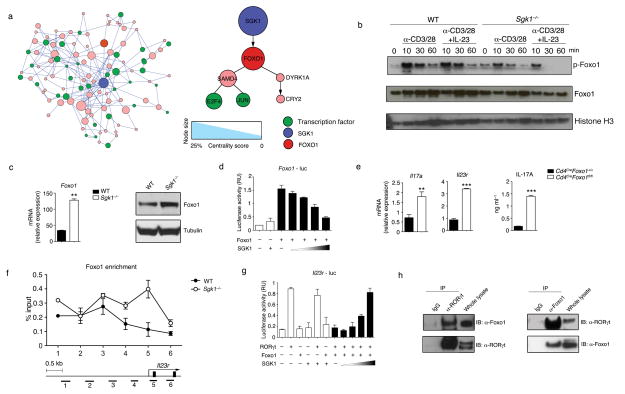
To better understand the molecular role of SGK1 in Th17 cells, we conducted another network analysis, using PPI data to connect SGK1 to the transcription factors whose activity is dysregulated in Sgk1−/− Th17 cells (Methods). The analysis suggested Foxo1 as one of the highest-ranking nodes downstream of SGK1 (Fig. 3a; Supplementary Table 1 and Supplementary Fig. 5a). Foxo1 phosphorylation by SGK1 has previously been shown in adipocytes to lead to its deactivation and translocation from the nucleus to the cytoplasm12. Consistent with this observation, we have observed that SGK1 phosphorylates Foxo1 (Supplementary Fig. 5b). By immunoblot of Sgk1−/− Th17 cells restimulated with IL-23, we confirmed that not only is there reduced phosphorylation of Foxo1 in the nucleus, but there was increased mRNA and protein expression of Foxo1 (Fig. 3b, c), suggesting that compromised phosphorylation of Foxo1 can result in its own transcriptional upregulation. It has been previously shown that Foxo1 can regulate its own expression13 and we have found that Foxo1 binds to a site located about 1 kilobase (kb) upstream of the first exon in the Foxo1 locus (Supplementary Fig. 6a). Transfection of a Foxo1 luciferase reporter in the presence of Foxo1 led to increased luciferase activity (Supplementary Fig. 6b), while increasing expression of SGK1 in the presence of Foxo1 resulted in a dose-dependent decrease in reporter activity, suggesting that SGK1 inhibited Foxo1-mediated transactivation of its own promoter (Fig. 3d).
Figure 3.
To decipher the consequences of Foxo1 expression on Th17 cell development, we used Foxo1−/− CD4+ memory T cells and observed increased expression of IL-23R and IL-17A compared to WT cells, indicating that Foxo1 may act as a repressor of Th17 cell development and of IL-23 signaling (Fig. 3e and Supplementary Fig. 6c). We also found potential binding sites of Foxo1 located about 1 kb upstream of the first exon of the Il23r locus by ChIP-PCR (Supplementary Fig. 6d). Moreover, there is significantly enriched binding of Foxo1 on the Il23r promoter region in Sgk1−/− cells compared to WT T cells, indicating enhanced suppression of Il23r transcription in the absence of SGK1 (Fig. 3f). RORγt has been suggested to be the master transcription factor of Th17 development and ChIP-seq28 and our ChIP-PCR analysis confirmed that IL-23R is one of the targets of RORγt (Supplementary Fig. 6e). Indeed, we observed that the Il23r promoter is transactivated by RORγt in IL-23-restimulated Th17 cells and it can be inhibited by Foxo1 in a dose-dependent manner (Supplementary Fig. 6f, g). While Foxo1 inhibited RORγt-mediated Il23r expression, co-expression of SGK1 together with RORγt and Foxo1 abrogated the suppressive effects of Foxo1 and rescued Il23r promoter transcriptional activity (Fig. 3g). Additionally, the inhibition of Il23r transcription by a phosphorylation-insensitive triple alanine mutant of Foxo1, Foxo1 AAA, was not reduced in the presence of SGK1 (Supplementary Fig. 6h). Furthermore, we observed an endogenous Foxo1-RORγt interaction in primary Th17 cells (Fig. 3h). These data support a model whereby some of the effects of SGK1 are due to phosphorylation of Foxo1, which may be a key step in relieving RORγt from Foxo1-mediated inhibition, enhancing the expression of IL-23R.
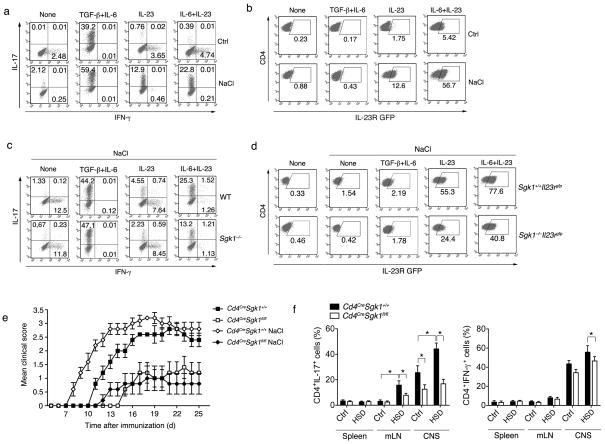
SGK1 has been reported to act as a mediator for sodium homeostasis. It can be induced by exogenous sodium chloride and is one of the major kinases that regulates Na+ intake by phosphorylation of epithelial sodium channels (ENaCs)4, 5. Considering the defects in Th17 development in Sgk1−/− mice, this raised the hypothesis that increasing sodium concentration may affect the Th17 cell phenotype through SGK1. To test this hypothesis, we first activated naïve T cells in the presence of additional NaCl, but in the absence of any polarizing cytokines. Microarray analysis of these NaCl-treated cells showed a significant upregulation of Sgk1 and of multiple other genes associated with Th17 development (Fisher exact test; p<10−3; Supplementary Table 2; Supplementary Fig. 7a), which we confirmed by qPCR analysis of selected genes (Supplementary Fig. 7b). We also observed increased mRNA and protein levels of IL-17 and IL-23R with additional NaCl under various Th17 polarizing conditions (Fig. 4a, b and Supplementary Fig. 7c). Furthermore, a sodium-induced increase in Th17 development and IL-23R expression was not observed in SGK1-deficient T cells, specifically in the context of IL-23-IL-23R signaling (Fig 4c, d). Importantly, culturing cells with mannitol did not alter Th17 cell differentiation, excluding the possibility that the Th17 program is initiated simply by the alteration of osmotic pressure (Supplementary Fig. 7d).
Figure 4.
Recent studies have shown that different components in the daily diet and gut microbiota can strongly affect the frequency of effector T cells in the gut14, 15. Additionally, previous data indicate that molecules related to sodium homeostasis can influence Th17 cell responses16, 17. To further understand the effect of NaCl on Th17 cell generation in vivo, we fed a high salt diet (HSD) to WT or Cd4CreSgk1fl/fl mice. After 3 weeks on HSD, we observed that unimmunized WT mice showed a marked increase in the frequency of Th17 cells in the lamina propria (LP), while no notable changes were observed in the mesenteric lymph nodes (mLN) or spleen. On the other hand, SGK1-deficient mice exhibited a much milder enhancement of Th17 cell frequency in the gut while there was no increase in IFN-γ production in any of the mice fed with HSD (Supplementary Fig. 8a, b).
Finally, we studied whether HSD would affect the development of Th17 and EAE in vivo. Mice fed HSD showed increased EAE severity when compared to the WT mice, which was dramatically reduced in SGK1-deficient mice (Fig. 4e and Supplementary Fig. 8c,d). We also observed a significantly higher frequency of Th17 cells in mLN and CNS of WT mice fed with HSD, but not in SGK1-deficient mice. The percentage of IFN-γ producing T cells in the CNS, but not in the peripheral immune compartments, of WT mice was increased in mice fed HSD, suggesting that HSD may increase infiltration, but not expansion of IFN-γ+ effector T cells, in the target organs (Fig. 4f and Supplementary Fig. 8e). Consistent with our in vitro data, we observed elevated IL-17 but not IFN-γ, production from CD4+ T cells isolated from EAE immunized WT mice fed with HSD and restimulated in vitro with MOG35–55 (Supplementary Fig. 8f). The data presented here indicates that high sodium intake potentiates Th17 cell generation in vivo in an SGK1-dependent manner and therefore has the potential to increase the risk of promoting autoimmunity.
In conclusion, we used a combination of microarray data analysis, large-scale protein-protein interaction network analysis and experimental data from multiple different knockout mice to establish IL-23R-SGK1-Foxo1 as a critical axis in Th17 stabilization. We show that Foxo1 acts as a repressor of IL-23R expression by directly binding to the Il23r promoter and inhibiting RORγt mediated Il23r transactivation. Phosphorylation of Foxo1, mediated by SGK1, leads to its deactivation and promotes unopposed RORγt-mediated Il23r transcription. SGK1 has been extensively studied in the context of NaCl transport18, 19. Modest increase of the NaCl concentration induces SGK1 expression in T cells with increased IL-23R expression and Th17 cell generation in vitro. Interestingly, even in unimmunized mice, enhancement of Th17 differentiation was observed in vivo in the gut and gut-associated lymphoid tissue and this increase in Th17 cells can be recalled at other peripheral sites following immunization. Although our data suggests an essential role for SGK1 in this process, it is likely that other immune cells and pathways are also influenced by increased salt intake. Furthermore, our result does not exclude additional alternative mechanisms by which an increase in NaCl affects Th17 cells. Nevertheless, the elevated in vivo Th17 differentiation by HSD raises the important issue of whether increased salt in the western diet and in processed foods is responsible for an increased generation of pathogenic Th17 cells and for an unprecedented increase in autoimmune diseases in the western world.
Methods Summary
Microarrays and network analysis
For gene expression analysis Affymetrix microarray chips were used. Data was processed using the GenePattern suite20. Differentially expressed genes were detected using fold-change and t-test analysis (for Sgk1−/− and NaCl-treated T cells), a consensus of fold-change, the EDGE software21 and a novel sigmoid-based method22 (for the Il23r−/− Th17 cell time course data). A command-line version of the ANAT software9 was used for network analysis.
In vitro T cell differentiation
Naïve T cells were FACS-sorted, stimulated with plate bound anti-CD3/CD28 and the indicated cytokines or NaCl and cells were analyzed by qPCR or flow cytometry at different time points.
EAE model
Mice were immunized subcutaneously with MOG35–55, CFA, heat-inactivated Mycobacterium tuberculosis and with intraperitoneal injection of Bordatella pertussis toxin.
In vivo cell transfer
Naïve T cells were differentiated towards Th17 cells, then transferred into MOG35–55/CFA immunized hosts and T cells isolated from various organs were analyzed by flow cytometry at 7–12 days after onset of EAE.
Western Blot/Immunoprecipitation
Differentiated T cells or transfected HEK293T cells were lysed in WCE buffer and lysates were subjected to Western blot or immunoprecipitation analysis.
Promoter activity reporter assay
HEK 293T cells were transfected with luciferase reporter constructs and expression vectors and luciferase expression was determined after 48 h.
Supplementary Material
Acknowledgments
We thank D. Kozoriz for cell sorting; Dr. Zhou. L, Dr. Accili. D, Dr. Demoulin. J and Dr. Sato. K provide reagents. Supported by the US National Institutes of Health (NS030843, NS045937, AI073748 and AI045757 to V.K.K., 1P01HG005062-01, 1P50HG006193-01 and DP1-OD003958-01 to A.R.; and K01DK090105 to S.X.); National MS Society (RG2571 to V.K.K.); the Howard Hughes Medical Institute (A.R.); the Klarman Cell Observatory; Guthy Jackson Foundation and the Austrian Science Fund (FWF, J 3091-B12 to T.T).
Footnotes
Authors’ contribution
C.W, N.Y. and T.T. performed experiments and wrote the manuscript. C.Z., S.X. and Y.K. performed experiments. N.Y. analyzed the data. A.R. and V.K.K. supervised the study and edited the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278(3):1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 4.Lang F, Bohmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev. 2006;86(4):1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- 5.Wulff P, Vallon V, Huang DY, Volkl H, Yu F, Richter K, et al. Impaired renal Na(+) retention in the sgk1-knockout mouse. The Journal of clinical investigation. 2002;110(9):1263–1268. doi: 10.1172/JCI15696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salker MS, Christian M, Steel JH, Nautiyal J, Lavery S, Trew G, et al. Deregulation of the serum-and glucocorticoid-inducible kinase SGK1 in the endometrium causes reproductive failure. Nature medicine. 2011;17(11):1509–1513. doi: 10.1038/nm.2498. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Cui R, Cheng X, Du J. Antiapoptotic effect of serum and glucocorticoid-inducible protein kinase is mediated by novel mechanism activating I{kappa}B kinase. Cancer Res. 2005;65 (2):457–464. [PubMed] [Google Scholar]
- 8.Shelly C, Herrera R. Activation of SGK1 by HGF, Rac1 and integrin-mediated cell adhesion in MDCK cells: PI-3K-dependent and -independent pathways. J Cell Sci. 2002;115(Pt 9):1985–1993. doi: 10.1242/jcs.115.9.1985. [DOI] [PubMed] [Google Scholar]
- 9.Yosef N, Zalckvar E, Rubinstein AD, Homilius M, Atias N, Vardi L, et al. ANAT: a tool for constructing and analyzing functional protein networks. Sci Signal. 2011;4(196):pl1. doi: 10.1126/scisignal.2001935. [DOI] [PubMed] [Google Scholar]
- 10.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21(3):952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Pietro N, Panel V, Hayes S, Bagattin A, Meruvu S, Pandolfi A, et al. Serum- and glucocorticoid-inducible kinase 1 (SGK1) regulates adipocyte differentiation via forkhead box O1. Mol Endocrinol. 2010;24(2):370–380. doi: 10.1210/me.2009-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284(16):10334–10342. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479(7374):538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 15.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10(3):260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stegbauer J, Lee DH, Seubert S, Ellrichmann G, Manzel A, Kvakan H, et al. Role of the renin-angiotensin system in autoimmune inflammation of the central nervous system. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(35):14942–14947. doi: 10.1073/pnas.0903602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrada AA, Contreras FJ, Marini NP, Amador CA, Gonzalez PA, Cortes CM, et al. Aldosterone promotes autoimmune damage by enhancing Th17-mediated immunity. J Immunol. 2010;184(1):191–202. doi: 10.4049/jimmunol.0802886. [DOI] [PubMed] [Google Scholar]
- 18.Diakov A, Korbmacher C. A novel pathway of epithelial sodium channel activation involves a serum- and glucocorticoid-inducible kinase consensus motif in the C terminus of the channel’s alpha-subunit. J Biol Chem. 2004;279(37):38134–38142. doi: 10.1074/jbc.M403260200. [DOI] [PubMed] [Google Scholar]
- 19.Loffing J, Zecevic M, Feraille E, Kaissling B, Asher C, Rossier BC, et al. Aldosterone induces rapid apical translocation of ENaC in early portion of renal collecting system: possible role of SGK. Am J Physiol Renal Physiol. 2001;280(4):F675–682. doi: 10.1152/ajprenal.2001.280.4.F675. [DOI] [PubMed] [Google Scholar]
- 20.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2. 0. Nat Genet. 2006;38 (5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 21.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chechik G, Koller D. Timing of gene expression responses to environmental changes. J Comput Biol. 2009;16(2):279–290. doi: 10.1089/cmb.2008.13TT. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.