Abstract
Many bifunctional alkylating agents and anticancer drugs exert their cytotoxicity by producing cross links between the two complementary strands of DNA, termed interstrand crosslinks (ICLs). This blocks the strand separating processes during DNA replication and transcription, which can lead to cell cycle arrest and apoptosis. Cells use multiple DNA repair systems to eliminate the ICLs. Concerted action of repair proteins involved in Nucleotide Excision Repair and Homologous Recombination pathways are suggested to play a key role in the ICL repair. However, recent studies indicate a possible role for Base Excision Repair (BER) in mediating the cytotoxicity of ICL inducing agents in mammalian cells. Elucidating the mechanism of BER mediated modulation of ICL repair would help in understanding the recognition and removal of ICLs and aid in the development of potential therapeutic agents. In this review, the influence of BER proteins on ICL DNA repair and the possible mechanisms of action are discussed.
Keywords: AP endonuclease, Base Excision Repair, BCNU, Cisplatin, DNA glycosylase, Interstrand crosslinks, Mitomycin C, Nitrogen mustard, Oxaliplatin, Psoralen, DNA Polymerase beta
Introduction
Cellular DNA is under constant threat from endogenous sources such as reactive oxygen species (ROS) and exogenous sources such as environmental oxidants, alkylating agents and anticancer drugs. The most common DNA lesions are base modifications such as alkylation, oxidation, loss of bases and single strand breaks. Complex and more toxic lesions include crosslinks and double strand breaks [1, 2]. Cells are endowed with the inherent capacity to respond to and eliminate these DNA lesions. The lesions are typically recognized and removed by various DNA repair pathways [3]. The base excision repair (BER) pathway as its name suggests is mainly involved in the excision of damaged bases from the DNA. It is considered as the predominant repair system in the protection of cells against a broad range of small base lesions resulting from oxidation, alkylation and deamination [4]. The BER pathway is a highly conserved, multistep process which requires the concerted action of several proteins [5]. It has been estimated that cells encounter ~10000 damaged bases per day, most of which are removed by BER [6–8].
The initiation of BER occurs by the action of DNA glycosylases which recognize alterations to the DNA bases and remove the altered bases by hydrolyzing the N-glycosidic bond. Once the damaged base is removed by a glycosylase, the resulting sugar-phosphate backbone without the base is called an apurinic/apyrimidinic (AP) site [9, 10]. AP endonuclease1 (APE1) cleaves the phosphate backbone resulting in a nick with a 3′ hydroxyl group and 5′ deoxyribose phosphate (dRP) residue. The oxidation/reduction state of this 5′ deoxyribose is a crucial factor in determining the subsequent downstream processing. If the dRP is not oxidized/reduced, this will lead to the activation of the short-patch BER pathway with the recruitment of DNA Polymerase β (Pol β). The dRP is cleaved by the lyase activity of Pol β and the one nucleotide gap is also filled by Pol β. The final nick is subsequently ligated by the DNA ligase III and XRCC1 complex [10]. If there is any change in the oxidative state of the dRP residue, this leads to the inhibition of the lyase activity of Pol β and activation of other polymerase activity resulting in strand displacement which leads to a 2–10 nucleotide flap intermediate, which is cleaved by FEN1 and joined by DNA Ligase-I [11]. The latter process is termed long-patch repair and requires the action of PCNA [10]. In addition to the oxidized state of the dRP residue, lesion specificity, protein-protein interaction and cell cycle status can also influence the specific choice of BER sub-pathways [12, 13]. The nucleotide incision pathway (NIR) is suggested to be the backup of the BER pathway where Ape1 incises the damaged DNA independent of glycosylase cleavage [14].
Recent studies indicate that BER proteins have broad substrate specificity and they interact with each other to catalyze the repair of DNA lesions [15, 16]. However, in the context of drug therapy, effective BER can render cells resistant to alkylating agents by repairing the DNA adducts that would otherwise be cytotoxic [17, 18]. For example, BER repairs the DNA lesions induced by alkylating agents such as methyl methane sulphonate (MMS) and temozolomide and over expression of BER proteins enhance resistance to these drugs [19, 20]. Therefore, several attempts have been made to target the BER proteins to increase cell sensitivity to alkylating agents [21, 22]. Generation of knock-out mice and identification of small molecule inhibitors of BER proteins have proven to be useful tools to dissect the mechanisms of drug resistance. Several small molecule inhibitors of APE1, Pol β and PARP were tested extensively for their ability to enhance the cytotoxicity of alkylating anticancer agents and some of them have been successful in clinical trials [23–28]. BER proteins interact with proteins from other DNA repair pathways and this cross-talk/co-ordination has implications for combination therapy targeting two DNA repair pathways simultaneously [29, 30].
Interstrand crosslinks (ICLs)
DNA interstrand crosslinks are formed between both strands of DNA and these covalent links are highly toxic to cells [31, 32]. It has been shown that it takes only a single ICL to kill repair-deficient bacteria and yeast, and about 40 ICLs to kill repair-deficient mammalian cells [33, 34]. The ICLs form an absolute block to metabolic processes such as DNA replication and transcription, trigger cell cycle arrest and apoptosis, ultimately resulting in cell death [35]. In addition, ICLs are shown to cause mutations and genomic instability [36, 37]. Certain endogenous and environmental agents form DNA ICLs and the most important class of ICLs are chemotherapeutic agents such as nitrogen mustards (eg. melphalan), nitrosureas (eg. BCNU), platinum agents (cisplatin, carboplatin, oxaliplatin, transplatin etc), mitomycin C and psoralen [38].
With continued exposure, cells develop strategies to eliminate these ICLs in order to survive [39]. However, enhanced repair of ICLs induced by chemotherapeutic agents in tumor cells is detrimental to the efficacy of the treatment [40–42]. Therefore, it is clinically important to elucidate the mechanism of elimination of the ICLs in order to develop strategies to overcome drug resistance. Because of its complexity, the repair of ICLs requires the concerted action of multiple DNA repair pathways [43]. It has been shown that nucleotide excision repair (NER) and homologous recombination (HR) as well as Fanconi Anemia (FA) proteins are involved in the repair of ICLs [44–47]. Translesion synthesis (TLS) can also occur across the ICLs where TLS polymerases bypass the processed (unhooked) ICL intermediates and the low fidelity of these lesion bypass polymerases increases mutations at the ICL site [36, 48]. The ICL repair events are shown to be both replication-dependent [49] and replication-independent [50]. The replication-dependent ICL repair occurs in S or G2 phase of the cell cycle [51, 52]. ICL repair is initiated by DNA replication fork collapse which activates signaling pathways for cell cycle arrest, to repair the DNA lesion [53]. When the damage is not repaired, the apoptotic signaling pathways are triggered to kill the cell [54]. Evidences also suggest that ICL repair occurs outside of S phase and does not require replication of DNA substrates [55, 56].
Several studies have shown that cells defective in DNA repair pathways such as NER and FA are hypersensitive to crosslinking agents, indicating the role of these pathways in the processing of ICLs [57, 58]. A recent model of ICL repair suggests that Mus81-Eme1 endonuclease cleaves 3′ of the ICL lesion on one strand and ERCC1-XPF cleaves 5′ of the lesion unhooking the crosslink [59, 60]. This can be repaired in a recombination-dependent manner using the undamaged sister chromatid [37, 61, 62]. When the undamaged template is not available (since ICLs affect both strands of DNA), translesion synthesis past the crosslink can play a role in the repair process. This recombination-independent repair is error-prone and mutagenic, and mainly occurs in non-dividing cells and in the G1 phase of dividing cells [63–65].
The ICLs distort the DNA double helix and distortion levels affect the recognition and repair of the ICLs. Each cross-linking agent forms different ICL DNA structures and therefore can influence the repair of these lesions [50]. For example, nitrogen mustard ICLs reside in the DNA major grove and do not affect hydrogen bonding of G-C, but ICLs induced by nitrosureas affect this base pair bonding [31, 66]. Psoralen ICLs create significant distortions to the DNA double helix, whereas mitomycin C ICLs are relatively non-distorting [67, 68]. Cisplatin ICLs bend and unwind the DNA significantly, where the cytosines adjacent to cross-linked guanines are flipped extrahelical and are exposed to the cellular environment [69–71]. Cisplatin analogues, oxaliplatin and transplatin also forms ICLs, but without extrahelical flipping of the bases [72, 73]. These differences between the crosslinks formed by ICL inducing agents have a significant impact on the way these adducts are recognized and repaired [74]. NER has been shown to be involved in the elimination of bulky DNA lesions. Studies by us and others have suggested that BER can play a role in the processing of bulky and structure distorting DNA lesions such as ICLs [75–77]. This review describes the possible role of the major BER proteins in the processing of ICLs in vitro and in vivo. Table 1 summarizes the list of BER proteins and their cytotoxic response to the ICL inducing agents.
Table 1.
BER proteins and their cytotoxic response to different ICL inducing agents.
| BER protein | ICL agent | Cells | Cytotoxicity |
|---|---|---|---|
|
| |||
| AAG | BCNU | Mouse ES cells | Sensitive (79, 80) |
| Mouse bone marrow cells | No effect (83) | ||
| Cervical cancer cells (HeLa) | Sensitive (85) | ||
| MMC | Mouse ES cells | Sensitive (79, 80) | |
| Psoralen | Mouse bone marrow cells | No effect (83) | |
| Mouse ES cells | Sensitive (81) | ||
| Nitrogen mustard | Mouse ES cells | No effect (80) | |
| Mouse neuronal cells | Resistance (84) | ||
|
| |||
| NEIL1 | Psoralen | Cervical cancer cells (HeLa) | Sensitive (76) |
|
| |||
| UNG | MMC | MEF | Sensitive (77) |
| Cisplatin | MEF | Resistant (77) | |
|
| |||
| NTH1 | Cisplatin | Breast cancer cells (MCF7) | Sensitive (88) |
| MMC | Breast cancer cells (MCF7) | No effect (88) | |
|
| |||
| OGG1 | Cisplatin | Hepatoma cells (HepG2) | Sensitive (89) |
|
| |||
| APE1 | Cisplatin | Lung cancer cells (A549) | Sensitive (100) |
| Melanoma cells (wm3211) | Sensitive (102) | ||
| CHO | No effect (105) | ||
| MMC | Cervical cancer cells (HeLa) | Sensitive (103) | |
| CHO cell | No effect (105) | ||
| BCNU | Glioma (SNB19) | Sensitive (95) | |
| CHO cell | Sensitive (104, 105) | ||
| Melphalan | CHO cell | No effect (104) | |
| Psoralen | Cervical cancer cells (HeLa) | Sensitive (76) | |
|
| |||
| FEN1 | Cisplatin | Glioblastoma (LN308) | Sensitive (113) |
| Neuroblastoma (SK-N-MC) | Sensitive (120) | ||
| Chicken DT40 | No effect (43) | ||
| MMC | MEF | Sensitive (114) | |
| Nitrogen mustard | Saccharomyces cerevisiae | Sensitive (115) | |
|
| |||
| Polβ | Cisplatin | Cervical cancer cells (HeLa) | Sensitive (130) |
| Ovarian cancer cells (SKOV-3) | Sensitive (130) | ||
| Breast cancer cells (MDAMB231) | Resistant (77) | ||
| MEF | Resistant (77, 139) | ||
| NIH3T3 | Sensitive (141) | ||
| Oxaliplatin | MEF | No effect (136, 137, 140) | |
| Breast cancer cells (MDAMB231) | No effect (77) | ||
| MEF | Sensitive (136) | ||
| MMC | MEF | No effect (77) | |
| Melphalan | MEF | Sensitive (77, 137) | |
| MEF | No effect (137) | ||
| NIH3T3 | No effect (141) | ||
Glycosylases
In BER, specific DNA glycosylases recognize corresponding damaged bases and cleave the N-glycosidic bond between abnormal bases and deoxyribose, leaving either an abasic site or a DNA single-strand break [78]. Several DNA glycosylases are identified in humans, which bind specifically to the modified base initiating the BER pathway. Cell survival assays display differential effects of glycosylases on the sensitivity of ICL inducing agents. 3-alkyladenine-DNA glycosylase (AAG), also called 3-methyladenine-DNA glycosylase (MPG) excises 3 methyladenine from DNA and it is the only alkyl specific DNA glycosylase present in mammalian cells to date. Studies show that Aag null mouse ES cells are sensitive to 1,3-bis(2-chloroethyl)-1-nitrosurea (BCNU), mitomycin C (MMC) and psoralen, but not to nitrogen mustards [79–81]. It was shown that AAG protects against BCNU- and MMC- induced apoptosis and chromosomal damage possibly through initiating the repair of the DNA adducts formed by these agents or oxidative DNA damage [80]. This indicates that a variety of damaged bases can be substrates for AAG. However, AAG has no incision activity on MMC and psoralen mono adducts or ICLs [80, 81], suggesting a non-enzymatic, structural role for AAG [75]. This may also be due to enhanced repair of these adducts by DNA repair pathways such as NER. Even though, bacterial and mammalian (human and rat) AAG excised mustard adducts [82], the absence of AAG in mouse did not show mustard hypersensitivity [80]. These observations collectively indicate species specificity and agent/drug specificity in AAG initiated BER. Further studies are warranted to explore the possibility of species specificity. Decreased and delayed formation of psoralen ICL-induced DNA double strand breaks in the absence of AAG indicates its possible role in ICL repair. However, the exact mechanism is not understood. It was suggested that AAG does not prevent conversion of psoralen mono adducts into ICLs. Instead, it might enhance ICL repair by binding to an ICL intermediate either in the early or later stages of ICL DNA processing [81]. The ICL inducing agents also form mono adducts and intrastrand adducts, thus making it difficult to infer the types of adducts repaired by AAG. Interestingly, while ES cells are hypersensitive to BCNU and MMC, bone marrow cells showed equal sensitivities [83] and neuronal cells showed low level resistance to nitrogen mustard [84] in the absence of AAG. In human cervical cancer cells, down regulation of AAG resulted in BCNU hypersensitivity [85]. It is important to note that AAG is over expressed in cervical neoplasia while neurons exhibit relatively lower levels of AAG. This indicates that lineage and cell type specificity, and expression levels of AAG can also influence crosslink sensitivity and the initiation of BER.
Neil1 is the human homologue of the E.coli oxidized base-specific DNA glycosylase/endonuclease VIII (Nei) and acts on oxidized, saturated, and ring-fragmented bases. Depletion of Neil1 in HeLa cells results in hypersensitivity to psoralen [76]. A recent study suggested that hypersensitivity of FA cells to ICL could be partly due to Neil1 deficiency and over expression of Neil1 in these cells enhanced resistance to ICL agents [86]. In mammalian cells, uracil residues in DNA are rapidly removed by uracil-DNA glycosylase (UNG). The expression of UNG is deregulated in cancer and it has been shown that cisplatin treatment up regulates UNG expression and activity in lung cancer cell lines [87]. We have shown that loss of UNG results in a cisplatin resistant phenotype and enhanced cisplatin ICL repair in UNG null mouse embryonic fibroblasts [77]. Knockdown of human NTH1, the bifunctional DNA glycosylase/apurinic/apyrimidinic lyase enhanced sensitivity to cisplatin, but had no effect on MMC cytotoxicity. Cisplatin, but not MMC induces oxidative stress resulting in the generation of oxidized bases which are substrates for NTH1[88]. Ape1 was shown to incise the abasic sites generated by the removal of uracil by UNG from cisplatin ICL DNA substrates [77]. It would be interesting to determine if bifunctional glycosylases with AP lyase activity could process cisplatin ICLs without the requirement of Ape1 activity. The glycosylase substrates if left unrepaired are less cytotoxic to cells, however, enhanced levels of glycosylases result in the accumulation of more toxic abasic sites and single-strand breaks which overwhelm the cells. Over expression of OGG1 (8 oxoguanine DNA glycosylase) in mitochondria increased the sensitivity of cancer cells to cisplatin treatment [89]. Extensive mitochondrial damage, increased production of intracellular free radicals and enhanced apoptosis contributed to the observed hypersensitivity. However, sensitivity of MMC, cisplatin or nitrogen mustards was not affected by MPG over expression [90, 91].
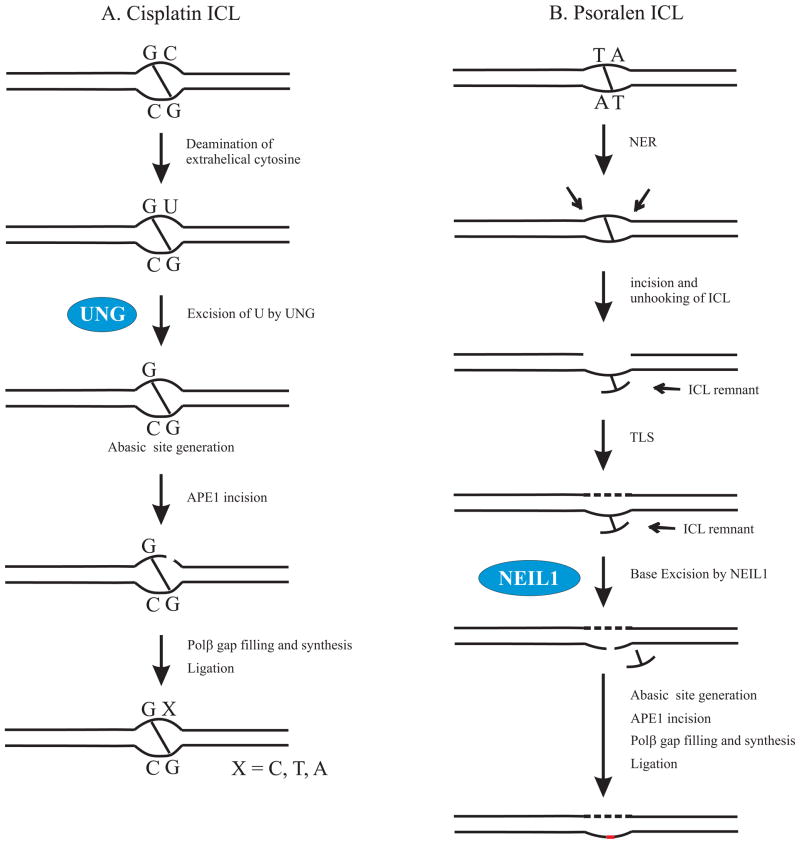
For decades, it was assumed that glycosylases excise only spontaneously formed, oxidized, and alkylated bases. However, recent studies indicate that ICLs formed between the two strands of DNA can also be substrates for the BER pathway. Couve-Privat and Couve et al have shown that Neil1 excises cross-linked thymine bases of the unhooked psoralen ICLs in vitro and this acts as a substrate for further BER processing. We have demonstrated that cisplatin ICLs with a flanking uracil base can be a substrate for the BER machinery [77]. UNG removed the uracil bases and the resulting AP site was cleaved by APE1. In addition, we have shown that depletion of UNG enhanced cisplatin ICL repair consistent with BER playing a positive role in mediating cisplatin cytotoxicity. Collectively, these studies suggest a possible role for DNA glycosylases in the processing of ICL DNA substrates as an alternate to the classic ICL-DNA repair pathways. The ICL substrate specificity and the capacity of other DNA repair systems to compensate for the loss or deregulation of BER can influence the cytotoxicity of an ICL inducing agent. Figure 1 depicts possible models for BER processing of cisplatin ICL and psoralen ICLs initiated by UNG and Neil1, respectively.
Figure 1.
Differential processing of ICLs by BER. A. BER processing of cisplatin ICL. The cytosines adjacent to the cisplatin ICL are extrahelical and flipped away from the double helix. The extrahelical cytosines are more susceptible to oxidative deamination and can convert to uracils. If cytosine deamination occurs, UNG initiates BER at the cisplatin ICL site by excising uracil from the DNA creating an abasic site. This is followed by Ape1 incision and Polβ mediated gap filling and synthesis. Polβ is shown to exhibit low fidelity at the cisplatin ICL site [77]. B. BER processing of psoralen ICL. NER may be involved in the initial recognition and incision of the psoralen ICLs by endonucleases. Translesion synthesis occurs across the unhooked ICLs where TLS polymerases bypass the ICL intermediate. NEIL1 initiates BER by excising the unhooked ICL remnant swinging out of the helix. Ape1 and Polβ complete the downstream BER processing [111].
Endonucleases
Ape1/Ref1 performs complex functions in cells through redox dependent and independent mechanisms. It acts as a transcriptional coactivator, regulates apoptosis, proliferation, differentiation, and production of ROS. In the BER pathway, it acts as an endonuclease that cleaves the abasic sites generated by the action of DNA glycosylases [92, 93]. Over expression of Ape1 has been observed in cancer cells and tumor samples which serves as a diagnostic and prognostic marker and also correlates with clinical outcome of chemotherapy [94–99]. High level expression of Ape1 has been shown to be associated with cisplatin resistance in head and neck and lung cancer [98, 100] but not in ovarian cancer [101]. Accordingly, down regulation of Ape1 enhanced cisplatin sensitivity in lung adenocarcinoma cells [100] and over expression protected melanoma cells from cisplatin-induced apoptosis [102]. This anti-apoptotic effect of Ape1 is suggested to be mediated by redox activation of cell survival signals as cisplatin induced apoptosis is mediated by ROS production. Targeting Ape1 also enhanced sensitivity to BCNU, MMC and psoralen in cancer cells and accumulation of unrepaired AP sites in the absence of Ape1 is likely responsible for the observed hypersensitivity [76, 95, 103]. However, expression of a dominant-negative form of Ape1 sensitized CHO cells to BCNU, but not to melphalan, cisplatin and MMC [104, 105]. It is not clear whether agent specificity or species specificity plays a role in Ape1 reactivity. Madhusudan et al have shown that CRT0044876, an inhibitor of Ape1 did not enhance cellular sensitivity to nitrogen mustard [25]. Methoxyamine, a small molecule inhibitor which binds to AP sites and inhibits Ape1 cleavage potentiated the cytotoxicity of BCNU and MMC due to accumulation of persistent AP sites and possible generation of DSBs [77, 106]. However, in our recent study, methoxyamine enhanced cisplatin resistance through enhanced repair of cisplatin ICLs in mouse embryonic fibroblasts and human cancer cells [77].
Ape1 is predominantly expressed in the nucleus and its over expression has been shown to result from increased cytoplasmic localization. Moore et al suggested that translocation of nuclear enzyme into cytoplasm could affect the nuclear DNA repair capacity which implies that sub-cellular compartmentalization contributes to the functions of Ape1 [97, 107–109]. It appears that both cytoplasmic and nuclear functions of Ape1 are essential as repair of AP sites also occur in the mitochondria. Consistent with this, studies indicate that both redox and DNA repair functions of Ape1are involved in mediating cytotoxicity to DNA damaging agents. It was suggested that independent of glycosylses, the NIR pathway can incise DNA lesions as a backup to the NER pathway [110]. Our recent study has shown that Ape1 can incise adjacent to a cisplatin ICL only if it contains an abasic site, which suggests that the glycosylase activity is required to create an abasic site flanking the cisplatin ICL for Ape1 cleavage [77]. Couve et al have shown that Ape1 together with Neil1 initiate an alternative repair pathway for bulky psoralen adducts [111]. These studies suggest a possible role for Ape1 in ICL DNA processing which is likely dependent on the specific ICL agent.
FEN1, a structure-specific endonuclease is involved in the long patch BER pathway that removes 5′ overhanging flaps and in the processing of 5′ ends of Okazaki fragments in lagging strand DNA synthesis. FEN1 has also been shown to facilitate HR by DSB processing [112]. Absence of FEN1 enhanced sensitivity to cisplatin, MMC and nitrogen mustard indicating a possible role for long patch BER in ICL processing, inhibition of DNA replication processing resulting in the inhibition of ICL repair or possibly involvement of FEN1 in HR processing of ICLs [113–115]. Over expression of FEN1 is observed in several cancers [116–119] and down regulation of FEN1 enhanced cisplatin sensitivity and apoptosis in glioma cells [113]. Mutations in FEN1 is associated with cancer and mouse embryonic fibroblasts harboring a E160D mutation identified in human cancer shows deficiency in nuclease activities and undergoes MMC-induced apoptosis [114]. Additionally, expression of nuclease deficient FEN1 enhanced cisplatin sensitivity compared to wt FEN1 suggesting the requirement of the nuclease activity to elicit resistance to cisplatin [120]. However, loss of FEN1 in chicken DT40 cells did not affect cisplatin sensitivity [43]. Yeast Rad27 mutant (homologue of human FEN1) did not affect DSB formation following nitrogen mustard treatment which shows that Rad27 is not involved in the repair of ICL-induced DSBs [115]. FEN1 is localized to nuclear repair foci in response to DNA damage. Cisplatin and MMC treatment results in recruitment of FEN1 to sites of arrested replication foci [120, 121]. FEN1 co-localizes with WRN at arrested replication forks following MMC treatment [122]. Cisplatin induced FEN1 localization is dependent on the BRCA1/RAD51 pathway as evident by the decreased FEN1 and Rad51 foci in BRCA1-deficient cells. This could be associated with a role of FEN1 in HR complexes following ICL treatment. ERCC1 and XPG are recruited to repair foci in FEN1 nuclease deficient cells indicating that NER could compensate for the FEN1 deficiency [120]. In addition to templates with a primer having an unannealed 5′-tail or flap structure, FEN1 also cleaves flaps containing small covalent adducts [123]. Even though FEN1 could cleave cisplatin GG-intrastrand adduct substrates [124], cisplatin ICLs prevented FEN1 cleavage [123]. This could be due to the structural distortion induced by the cisplatin ICL, since it has been shown that DNA secondary structures inhibit flap processing. These observations raise the possibility that hypersensitivity of ICL inducing agents during the loss of FEN1 could be due to a role in DNA replication dependent ICL processing as ICLs are known to stall replication forks, due to its role in flap processing in long-patch BER or possibly due to a role for FEN1 in HR ICL processing.
Polymerases
Polymerase β(Pol β) belongs to the X family DNA polymerases and is well characterized for a role in DNA repair [125]. Studies have established the role of Pol β in both short-patch and long-patch BER [126]. Pol β has dRP lyase activity which is shown to be a rate-limiting step in the BER pathway [127]. This gap filling polymerase has been identified as being error prone which is evident by increased mutagenesis when it is over expressed. Over-expression of Pol β leads to bifunctional DNA damage tolerance and facilitates the error-prone translesion synthesis past the DNA lesions that otherwise would block DNA replication and kill the cells. This affects the genomic stability of the cells and influences tumorigenesis [128]. We recently demonstrated additional base incorporation at cisplatin ICL sites that resulted from strand displacement synthesis by Pol β [77]. We also showed that Pol β had a low fidelity at the site flanking the cisplatin ICL even in the presence of correct nucleotides.
Using matched normal and tumor samples from different tissues, Srivastava et al have shown that approximately one third of tumor samples over express Pol β [129]. However, down regulation of this enzyme was observed in some tumors [130]. Pol β gene sequencing in different cancer cells revealed different types of mutations [131]. Sweasy et al studied the transformation activity of Pol β variants which when expressed in mouse cells resulted in cellular transformation [132–134]. Studies have shown that targeting Pol β modulates sensitivity of cisplatin, oxaliplatin, MMC and melphalan [77, 135–138]. Pol β deficiency resulted in MMC hypersensitivity and enhanced apoptosis [137]. However, evidence for the effect of down regulation of Pol β in modulating cisplatin and oxaliplatin chemosensitivity is conflicting. Yang et al have shown that knock down or down regulation of Pol β in colon cancer cells and MEFs enhanced oxaliplatin sensitivity and this was attributed to the delayed repair of oxaliplatin intrastrand adducts as well as ICLs, and increased apoptosis [136]. In contrast to this, down regulation or loss of Pol β in breast cancer cells and MEFs did not affect oxaliplatin sensitivity as well as oxaliplatin ICL repair in our study [77]. Similarly, studies show that cisplatin cytotoxicity when targeting Pol β in MEFs as well as cancer cells resulted in a) hypersensitivity, b) resistance and c) no effect [130, 135, 139–141]. A Pol β inhibitor conferred resistance to cisplatin in MEFs, but not in ovarian cancer cells [142]. In our study, we have shown that down regulation of Pol β plays a positive role in cisplatin cytotoxicity and its deficiency resulted in a resistant phenotype through enhanced repair of cisplatin-ICLs [77]. The discrepancy in these observations of drug cytotoxicity upon deficiency of Pol β is not clear. However, mutations in the Pol β gene, basal level expression of Pol β and overall DNA repair capacity could influence a cells response to the crosslinking agents. We have shown that BER machinery processes the flanking DNA at the cisplatin ICL site resulting in the non-productive repair of the ICL, therefore competing with the productive ICL DNA repair mechanisms. We have also shown that Pol β has low fidelity at the cisplatin ICL site [77] and Pol β mediated misincorporation of nucleotides could generate mismatched bases initiating the mismatch repair (MMR) apparatus (Manuscript in preparation).
Polymerase δ and ε, components of the replication complex are important replication polymerases involved in the synthesis process in both NER and long-patch BER. These polymerases can also bypass across different types of DNA lesions indicating a possible role in translesion replication [143, 144]. Aphidicolin, which selectively binds to the replication polymerases and inhibits DNA synthesis, has been used in cancer chemotherapy. In vitro and in vivo studies have shown that aphidicolin potentiates cisplatin cytotoxicity in tumor samples and cancer cells [145–148]. The modulatory effect of aphidocolin was significantly higher in cisplatin-resistant cells implying that inhibition of these polymerases can overcome platinum resistance. Aphidicolin also increased BCNU and melphalan activity in cancer cells [138, 149]. Zhang et al have shown that these polymerases are involved in psoralen ICL processing, at least in the late stages [150]. Aphidicolin mediated inhibition of DNA synthesis, replication stress and cell cycle arrest are potential mechanisms for cytoxicity of the crosslinking agents. Since these replication polymerases are components of several DNA repair pathways including NER, MMR, HR as well as long-patch BER, the mechanism that aphidicolin utilizes to exert its ICL cytotoxic enhancement is unclear.
Other BER proteins
BER proteins including XRCC1 and PARP are also involved in processing ICLs. XRCC1 is involved in the repair of single strand breaks (SSB) generated during BER and acts as a scaffold, connecting other BER proteins such as PARP, Pol β and DNA ligase III [151]. Polymorphisms in the XRCC1 gene have been associated with increased risk of developing certain cancers and also used as a prognostic marker in platinum-treated lung and gastric cancer patients [151–153]. Down regulation of XRCC1 enhanced cellular sensitivity to cisplatin and mitomycin C [154, 155]. PARP plays a key role in the repair of single-stranded breaks (SSBs) via BER/SSB repair pathways apart from other cellular functions. Cells with PTEN and BRCA mutations were found to be sensitive to PARP inhibitors and some of the PARP inhibitors are in clinical use for the treatment of melanoma, breast, ovarian and colorectal cancers [27, 28]. The role of these proteins in ICL repair is further strengthened by the findings of Zhu and Lippard where XRCC1, DNA ligase III and PARP1 bind to cisplatin ICLs in vitro [156].
Conclusion
ICLs covalently link the two strands of DNA and block the denaturing cellular processes that occur during DNA replication and transcription. ICLs are cytotoxic DNA lesions that are formed by a variety of anticancer drugs such as cisplatin, mitomycin C, psoralen, nitrosureas and nitrogen mustard derivatives. These crosslinking agents distort the DNA double helix, each in a unique manner. The repair of ICLs is still not completely understood in eukaryotes and the participation of different DNA repair pathways in ICL DNA repair is important. Recent evidence indicates that the BER pathway may play a novel role in the repair and processing of ICLs. Several mechanisms have been proposed on the role of BER in mediating ICL repair or processing. Accumulating evidence indicates that a variety of DNA lesions including ICLs can serve as substrates for the BER machinery. However, differential cellular sensitivity towards the ICL inducing agents suggests that the type of lesions formed determines the BER processing event and the effect is not general, but agent specific. The ICLs formed by each agent bend and distort the DNA in a unique manner leading to differential protein recognition and the distortion levels have a significant impact in the way these adducts are recognized and repaired. Each ICL structure therefore, has the potential to be processed differently and these unique physical structures contribute to the initiation of BER. The ICL inducing agents also form monoadducts and intrastrand adducts, which could also be substrates of BER. Some types of mono adducts can convert to ICLs and by repairing monoadducts, BER could prevent ICL formation and thereby, modulate the cellular responses to these agents. However, in vitro studies demonstrating differential recognition and processing of monoadducts by different glycosylases implies the existence of an alternate mechanism. In many cases, BER processing also depends on cell and tissue specificity as well as normal vs cancer cells based on the relative levels of basal protein expression and activity. Additionally, inherent DNA repair capacity also affects cellular responses to ICLs as it was suggested that BER may be able to compensate for the loss of other DNA repair pathways. BER could facilitate the repair of ICLs by interacting with other DNA repair pathways or compete with them to inhibit or delay the repair processes. BER mediated ICL processing could be productive as well as non-productive depending on the specific cross linking agents. BER can also act as an alternative or back-up pathway to NER in ICL repair as seen with psoralen. Collectively, it can be concluded that BER proteins process each ICL differently, mostly based on the distinct DNA structural distortions generated by the ICL inducing agents.
Highlights.
BER pathway is mainly involved in the excision of damaged bases from the DNA
BER is also involved in the processing of bulky lesions and interstrand crosslinks
BER proteins process ICLs, mostly based on the distinct DNA structural distortions
Acknowledgments
We thank members of the Patrick lab for critical reading of the manuscript. This study was supported by a grant from the National Institutes of Health (1R01-GM088249) to SMP.
Footnotes
Conflict of interest
None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair: From molecular mechanisms to health implications. Antioxidants & Redox Signaling. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- 2.Reddy MC, Vasquez KM. Repair of genome destabilizing lesions. Radiation Research. 2005;164:345–356. doi: 10.1667/rr3419.1. [DOI] [PubMed] [Google Scholar]
- 3.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes & Development. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 4.Zharkov DO. Base excision DNA repair. Cellular and Molecular Life Sciences. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson DM, Thompson LH. Life without DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baute J, Depicker A. Base excision repair and its role in maintaining genome stability. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:239–276. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- 7.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Research. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. Journal of Biological Chemistry. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 11.Podlutsky AJ, Dianova II, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. Embo Journal. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortini P, Dogliotti E. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. Dna Repair. 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Dogliotti E, Fortini P, Pascucci B, Parlanti E. The mechanism of switching among multiple BER pathways. Progress in Nucleic Acid Research and Molecular Biology. 2001;68:3–27. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- 14.Timofeyeva NA, Koval VV, Knorre DG, Zharkov DO, Saparbaev MK, Ishchenko AA, Fedorova OS. Conformational Dynamics of Human AP Endonuclease in Base Excision and Nucleotide Incision Repair Pathways. Journal of Biomolecular Structure & Dynamics. 2009;26:637–652. doi: 10.1080/07391102.2009.10507278. [DOI] [PubMed] [Google Scholar]
- 15.Bennett RAO, Wilson DM, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human B-oxoguanine-DNA glycosylase and AP endonuclease. Dna Repair. 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Kaina B, Christmann M. DNA repair in resistance to alkylating anticancer drugs. International Journal of Clinical Pharmacology and Therapeutics. 2002;40:354–367. doi: 10.5414/cpp40354. [DOI] [PubMed] [Google Scholar]
- 18.Damia G, D’Incalci M. Mechanisms of resistance to alkylating agents. Cytotechnology. 1998;27:165–173. doi: 10.1023/A:1008060720608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobol RW, Watson DE, Nakamura J, Yakes FM, Hou E, Horton JK, Ladapo J, Van Houten B, Swenberg JA, Tindall KR, Samson LD, Wilson SH. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Research. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 21.Glazer PM, Stachelek GC, Dalal S, Sweasy JB. Targeting Base Excision Repair to Potentiate Cancer Therapy. International Journal of Radiation Oncology Biology Physics. 2009;75:S21. [Google Scholar]
- 22.Sharma RA, Dianov GL. Targeting base excision repair to improve cancer therapies. Molecular Aspects of Medicine. 2007;28:345–374. doi: 10.1016/j.mam.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Wilson DM, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cellular and Molecular Life Sciences. 2010;67:3621–3631. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed AM, Fishel ML, Kelley MR. Small-molecule inhibitors of proteins involved in base excision repair potentiate the anti-tumorigenic effect of existing chemotherapeutics and irradiation. Future Oncology. 2009;5:713–726. doi: 10.2217/FON.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJE, Dianov GL, Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Research. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu HY, Horton JK, Gryk MR, Prasad R, Naron JM, Sun DA, Hecht SM, Wilson SH, Mullen GP. Identification of small molecule synthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. Journal of Biological Chemistry. 2004;279:39736–39744. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- 27.Fong PC, Boss DS, Yap TA, Tutt A, Wu PJ, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JHM, de Bono JS. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. New England Journal of Medicine. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee S, Kaye SB, Ashworth A. Making the best of PARP inhibitors in ovarian cancer. Nature Reviews Clinical Oncology. 2010;7:508–519. doi: 10.1038/nrclinonc.2010.116. [DOI] [PubMed] [Google Scholar]
- 29.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. Dna Repair. 2007;6:517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA Polymerases as Potential Therapeutic Targets for Cancers Deficient in the DNA Mismatch Repair Proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guainazzi A, Scharer OD. Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cellular and Molecular Life Sciences. 2010;67:3683–3697. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dronkert MLG, Kanaar R. Repair of DNA interstrand cross-links. Mutation Research-Dna Repair. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 33.Maganaschwencke N, Henriques JAP, Chanet R, Moustacchi E. The Fate of 8-Methoxypsoralen Photoinduced Crosslinks in Nuclear and Mitochondrial Yeast Dna - Comparison of Wild-Type and Repair-Deficient Strains. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 35.Mcvey M. Strategies for DNA Interstrand Crosslink Repair: Insights From Worms, Flies, Frogs, and Slime Molds. Environmental and Molecular Mutagenesis. 2010;51:646–658. doi: 10.1002/em.20551. [DOI] [PubMed] [Google Scholar]
- 36.Shen X, Li L. Mutagenic Repair of DNA Interstrand Crosslinks. Environmental and Molecular Mutagenesis. 2010;51:493–499. doi: 10.1002/em.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonnalagadda VS, Matsuguchi T, Engelward BP. Interstrand crosslink-induced homologous recombination carries an increased risk of deletions and insertions. Dna Repair. 2005;4:594–605. doi: 10.1016/j.dnarep.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Scharer OD. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 39.Vasquez KM. Targeting and Processing of Site-Specific DNA Interstrand Crosslinks. Environmental and Molecular Mutagenesis. 2010;51:527–539. doi: 10.1002/em.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panasci L, Xu ZY, Bello V, Aloyz R. The role of DNA repair in nitrogen mustard drug resistance. Anti-Cancer Drugs. 2002;13:211–220. doi: 10.1097/00001813-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Wynne P, Newton C, Ledermann J, Olaitan A, Mould TA, Hartley JA. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. British Journal of Cancer. 2007;97:927–933. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spanswick VJ, Craddock C, Sekhar M, Mahendra P, Shankaranarayana P, Hughes RG, Hochhauser D, Hartley JA. Repair of DNA interstrand crosslinks as a mechanism of clinical resistance to melphalan in multiple myeloma. Blood. 2002;100:224–229. doi: 10.1182/blood.v100.1.224. [DOI] [PubMed] [Google Scholar]
- 43.Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Research. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 44.Wood RD. Mammalian Nucleotide Excision Repair Proteins and Interstrand Crosslink Repair. Environmental and Molecular Mutagenesis. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinz JM. Role of Homologous Recombination in DNA Interstrand Crosslink Repair. Environmental and Molecular Mutagenesis. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 46.Hlavin EM, Smeaton MB, Miller PS. Initiation of DNA Interstrand Cross-link Repair in Mammalian Cells. Environmental and Molecular Mutagenesis. 2010;51:604–624. doi: 10.1002/em.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho V, Scharer OD. Translesion DNA Synthesis Polymerases in DNA Interstrand Crosslink Repair. Environmental and Molecular Mutagenesis. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 49.Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun JC, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. Cross-Link Structure Affects Replication-Independent DNA Interstrand Cross-Link Repair in Mammalian Cells. Biochemistry. 2010;49:3977–3988. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Legerski RJ. Repair of DNA Interstrand Cross-links During S Phase of the Mammalian Cell Cycle. Environmental and Molecular Mutagenesis. 2010;51:540–551. doi: 10.1002/em.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mladenova V, Russev G. Enhanced repair of DNA interstrand crosslinks in S phase. Febs Letters. 2006;580:1631–1634. doi: 10.1016/j.febslet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, Hinz JM, Helleday T. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Research. 2009;37:6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasquez KM, Legerski RJ. DNA Interstrand Crosslinks: Repair, Cell Signaling, and Therapeutic Implications. Environmental and Molecular Mutagenesis. 2010;51:491–492. doi: 10.1002/em.20598. [DOI] [PubMed] [Google Scholar]
- 55.Sarkar S, Davies AA, Ulrich HD, Mchugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo Journal. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of Laser-localized DNA Interstrand Cross-links in G(1) Phase Mammalian Cells. Journal of Biological Chemistry. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clingen PH, Arlett CF, Hartley JA, Parris CN. Chemosensitivity of primary human fibroblasts with defective unhooking of DNA interstrand cross-links. Experimental Cell Research. 2007;313:753–760. doi: 10.1016/j.yexcr.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Andreassen PR, Ren KQ. Fanconi Anemia Proteins, DNA Interstrand Crosslink Repair Pathways, and Cancer Therapy. Current Cancer Drug Targets. 2009;9:101–117. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NGJ, Beverloo HB, Hoeijmakers JHJ, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Molecular and Cellular Biology. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo Journal. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. Journal of Biological Chemistry. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 62.Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenetic and Genome Research. 2004;104:28–34. doi: 10.1159/000077463. [DOI] [PubMed] [Google Scholar]
- 63.Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) Journal of Biological Chemistry. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Peterson CA, Zheng HY, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Molecular and Cellular Biology. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng HY, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Molecular and Cellular Biology. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rink SM, Solomon MS, Taylor MJ, Rajur SB, Mclaughlin LW, Hopkins PB. Covalent Structure of A Nitrogen Mustard-Induced Dna Interstrand Cross-Link - An N7-To-N7 Linkage of Deoxyguanosine Residues at the Duplex Sequence 5′-D(Gnc) Journal of the American Chemical Society. 1993;115:2551–2557. [Google Scholar]
- 67.Kumaresan KR, Ramaswamy M, Yeung AT. Structure of the Dna Interstrand Cross-Link of 4,5′,8-Trimethylpsoralen. Biochemistry. 1992;31:6774–6783. doi: 10.1021/bi00144a018. [DOI] [PubMed] [Google Scholar]
- 68.Fagan PA, Spielmann HP, Sigurdsson ST, Rink SM, Hopkins PB, Wemmer DE. An NMR study of [d(CGCGAATTCGCG)](2) containing an interstrand cross-link derived from a distamycin-pyrrole conjugate. Nucleic Acids Research. 1996;24:1566–1573. doi: 10.1093/nar/24.8.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 angstrom resolution: hydration at the platinated site. Nucleic Acids Research. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang HF, Zhu LM, Reid BR, Drobny GP, Hopkins PB. Solution Structure of A Cisplatin-Induced Dna Interstrand Cross-Link. Science. 1995;270:1842–1845. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- 71.Lukin M, de los Santos C. NMR structures of damaged DNA. Chemical Reviews. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 72.Kasparkova J, Vojtiskova M, Natile G, Brabec V. Unique properties of DNA interstrand cross-links of antitumor oxaliplatin and the effect of chirality of the carrier ligand. Chemistry-A European Journal. 2008;14:1330–1341. doi: 10.1002/chem.200701352. [DOI] [PubMed] [Google Scholar]
- 73.Brabec V, Sip M, Leng M. Dna Conformational Change Produced by the Site-Specific Interstrand Cross-Link of Trans-Diamminedichloroplatinum(Ii) Biochemistry. 1993;32:11676–11681. doi: 10.1021/bi00094a025. [DOI] [PubMed] [Google Scholar]
- 74.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 75.Wilson DM, Seidman MM. A novel link to base excision repair? Trends in Biochemical Sciences. 2010;35:247–252. doi: 10.1016/j.tibs.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Research. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kothandapani A, Dangeti VS, Brown AR, Banze LA, Wang XH, Sobol RW, Patrick SM. Novel role of base excision repair (BER) in mediating cisplatin cytotoxicity. Journal of Biological Chemistry. 2011;286:14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: Glycosylase mechanisms and structures. Annual Review of Biochemistry. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 79.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. Embo Journal. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 80.Allan JM, Engelward BP, Dreslin AJ, Wyatt MD, Tomasz M, Samson LD. Mammalian 3-methyladenine DNA glycosylase protects against the toxicity and clastogenicity of certain chemotherapeutic DNA cross-linking agents. Cancer Research. 1998;58:3965–3973. [PubMed] [Google Scholar]
- 81.Maor-Shoshani A, Meira LB, Yang XM, Samson LD. 3-methyladenine DNA glycosylase ill important for cellular resistance to psoralen interstrand cross-links. Dna Repair. 2008;7:1399–1406. doi: 10.1016/j.dnarep.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mattes WB, Lee CS, Laval J, OConnor TR. Excision of DNA adducts of nitrogen mustards by bacterial and mammalian 3-methyladenine-DNA glycosylases. Carcinogenesis. 1996;17:643–648. doi: 10.1093/carcin/17.4.643. [DOI] [PubMed] [Google Scholar]
- 83.Roth RB, Samson LD. 3-methyladenine DNA glycosylase-deficient Aag null mice display unexpected bone marrow alkylation resistance. Cancer Research. 2002;62:656–660. [PubMed] [Google Scholar]
- 84.Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, Samson LD, Gerson SL, Turker MS. DNA repair modulates the vulnerability of the developing brain to alkylating agents. Dna Repair. 2009;8:400–412. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paik J, Duncan T, Lindahl T, Sedgwick B. Sensitization of human carcinoma cells to alkylating agents by small interfering RNA suppression of 3-alkyladenine-DNA glycosylase. Cancer Research. 2005;65:10472–10477. doi: 10.1158/0008-5472.CAN-05-1495. [DOI] [PubMed] [Google Scholar]
- 86.Mace-Aime G, Couve S, Khassenov B, Rosselli F, Saparbaev MK. The Fanconi Anemia Pathway Promotes DNA Glycosylase-Dependent Excision of Interstrand DNA Crosslinks. Environmental and Molecular Mutagenesis. 2010;51:508–519. doi: 10.1002/em.20548. [DOI] [PubMed] [Google Scholar]
- 87.Liu B, Yang XH, Wang KM, Tan WH, Li HM, Tang HX. Real-time monitoring of uracil removal by uracil-DNA glycosylase using fluorescent resonance energy transfer probes. Analytical Biochemistry. 2007;366:237–243. doi: 10.1016/j.ab.2007.04.049. [DOI] [PubMed] [Google Scholar]
- 88.Guay D, Garand C, Reddy S, Schmutte C, Lebel M. The human endonuclease III enzyme is a relevant target to potentiate cisplatin cytotoxicity in Y-box-binding protein-1 overexpressing tumor cells. Cancer Science. 2008;99:762–769. doi: 10.1111/j.1349-7006.2008.00739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang HH, Mizumachi T, Carcel-Trullols J, Li LW, Naito A, Spencer HJ, Spring PM, Smoller BR, Watson AJ, Margison GP, Higuchi M, Fan CY. Targeting human 8-oxoguanine DNA glycosylase (hOGG1) to mitochondria enhances cisplatin cytotoxicity in hepatoma cells. Carcinogenesis. 2007;28:1629–1637. doi: 10.1093/carcin/bgm072. [DOI] [PubMed] [Google Scholar]
- 90.Grombacher T, Tomicic M, Digweed M, Kaina B. Overexpression of cDNA encoding FANCC, SPHAR, MPG, SNM1 or HA 3611 does not render CHO cells more resistant to DNA crosslinking agents. Anticancer Research. 1999;19:1729–1735. [PubMed] [Google Scholar]
- 91.Rinne ML, He Y, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Research. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): From mechanistic insights to druggable target in cancer. Cancer Treatment Reviews. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxidants & Redox Signaling. 2009;11:601–619. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clinical Cancer Research. 2001;7:3510–3518. [PubMed] [Google Scholar]
- 95.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kolstoe DD. The apurinic/apyrimidinic endonuclease activity of Apel/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress (vol 8, pg 3008, 2002) Clinical Cancer Research. 2003;9:2877. [PubMed] [Google Scholar]
- 96.Kelley MR, Cheng L, Foster R, Tritt R, Jiang JZ, Broshears J, Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clinical Cancer Research. 2001;7:824–830. [PubMed] [Google Scholar]
- 97.Kakolyris S, Kaklamanis L, Engels K, Turley H, Hickson ID, Gatter KC, Harris AL. Human apurinic endonuclease 1 expression in a colorectal adenoma-carcinoma sequence. Cancer Research. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 98.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/REF-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. International Journal of Radiation Oncology Biology Physics. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 99.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, Kelley MR. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Research. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 100.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 101.Freitas S, Moore DH, Michael H, Kelley MR. Studies of apurinic/apyrimidinic endonuclease/ref-1 expression in epithelial ovarian cancer: Correlations with tumor progression and platinum resistance. Clinical Cancer Research. 2003;9:4689–4694. [PubMed] [Google Scholar]
- 102.Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Molecular Cancer Therapeutics. 2005;4:1923–1935. doi: 10.1158/1535-7163.MCT-05-0229. [DOI] [PubMed] [Google Scholar]
- 103.Walker LJ, Craig RB, Harris AL, Hickson ID. A Role for the Human Dna-Repair Enzyme Hap1 in Cellular-Protection Against Dna-Damaging Agents and Hypoxic Stress. Nucleic Acids Research. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM. Impairment of APE1 Function Enhances Cellular Sensitivity to Clinically Relevant Alkylators and Antimetabolites. Molecular Cancer Research. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McNeill DR, Wilson DM. A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Molecular Cancer Research. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 106.Liu LL, Yan L, Donze JR, Gerson SL. Blockage of abasic site repair enhances antitumor efficacy of 1,3-bis-(2-chloroethyl)-1-nitrosourea in colon tumor xenografts. Molecular Cancer Therapeutics. 2003;2:1061–1066. [PubMed] [Google Scholar]
- 107.Moore DH, Michael H, Tritt R, Parsons SH, Kelley MR. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clinical Cancer Research. 2000;6:602–609. [PubMed] [Google Scholar]
- 108.Xu Y, Moore DH, Broshears J, Liu LF, Wilson TM, Kelley MR. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Research. 1997;17:3713–3719. [PubMed] [Google Scholar]
- 109.Wang D, Luo MH, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: Enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Molecular Cancer Therapeutics. 2004;3:679–686. [PubMed] [Google Scholar]
- 110.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 111.Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The Human Oxidative DNA Glycosylase NEIL1 Excises Psoralen-induced Interstrand DNA Cross-links in a Three-stranded DNA Structure. Journal of Biological Chemistry. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kikuchi K, Taniguchi Y, Hatanaka A, Sonoda E, Hochegger H, Adachi N, Matsuzaki Y, Koyama H, van Gent DC, Jasin M, Takeda S. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Molecular and Cellular Biology. 2005;25:6948–6955. doi: 10.1128/MCB.25.16.6948-6955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nikolova T, Christmann M, Kaina B. FEN1 is Overexpressed in Testis, Lung and Brain Tumors. Anticancer Research. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 114.Zheng L, Dai HF, Zhou M, Li M, Singh P, Qiu JZ, Tsark W, Huang Q, Kernstine K, Zhang XM, Lin DX, Shen BH. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nature Medicine. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 115.Mchugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2000;20:3425–3433. doi: 10.1128/mcb.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene. 2003;22:7243–7246. doi: 10.1038/sj.onc.1206977. [DOI] [PubMed] [Google Scholar]
- 117.Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, Kim JG, Kim NS. Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clinical Cancer Research. 2005;11:473–482. [PubMed] [Google Scholar]
- 118.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, Van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. American Journal of Pathology. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, Zeng G, Horvath S, Belldegrun AS. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. Bju International. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- 120.Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Molecular and Cellular Biology. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sharma S, Sommers JA, Gary RK, Friedrich-Heineken E, Hubscher U, Brosh RM. The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Research. 2005;33:6769–6781. doi: 10.1093/nar/gki1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM. WRN helicase and FEN-1 form a complex upon replication arrest and together process branch-migrating DNA structures associated with the replication fork. Molecular Biology of the Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 124.Barnes CJ, Wahl AF, Shen BH, Park MS, Bambara RA. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′- to 3′-exonuclease endonuclease RAD2 homologue 1 or flap endonuclease 1. Journal of Biological Chemistry. 1996;271:29624–29631. doi: 10.1074/jbc.271.47.29624. [DOI] [PubMed] [Google Scholar]
- 125.Yamtich J, Sweasy JB. DNA polymerase Family X: Function, structure, and cellular roles. Biochimica et Biophysica Acta-Proteins and Proteomics. 2010;1804:1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 127.Allinson SL, Dianova II, Dianov GL. DNA polymerase beta is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. Embo Journal. 2001;20:6919–6926. doi: 10.1093/emboj/20.23.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Canitrot Y, Frechet M, Servant L, Cazaux C, Hoffmann JB. Overexpression of DNA polymerase beta: a genomic instability enhancer process. Faseb Journal. 1999;13:1107–1111. doi: 10.1096/fasebj.13.9.1107. [DOI] [PubMed] [Google Scholar]
- 129.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 130.Albertella MR, Lau A, O’Connor MJ. The overexpression of specialized DNA polymerases in cancer. Dna Repair. 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 131.Sweasy JB, DiMaio D, Lang T, Dalai S, Maitra M, Starcevic D, Chikova A. Cancer-associated variants of DNA polymerase beta. Environmental and Molecular Mutagenesis. 2007;48:548. [Google Scholar]
- 132.Lang TM, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Molecular and Cellular Biology. 2007;27:5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sweasy JB, Dalal S, Lang T. A gastric cancer variant of DNA polymerase beta induces cellular transformation by a mutational mechanism. Environmental and Molecular Mutagenesis. 2006;47:415. [Google Scholar]
- 134.Sweasy JB, Lang T, Starcevic D, Sunt KW, Lai CC, DiMaio D, Dalal S. Expression of DNA polymerase beta cancer-associated variants in mouse cells results in cellular transformation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Raaphorst GP, Cybulski SE, Sobol R, Ng CE. The response of human breast tumour cell lines with altered polymerase beta levels to cisplatin and radiation. Anticancer Research. 2001;21:2079–2083. [PubMed] [Google Scholar]
- 136.Yang J, Parsons J, Nicolay NH, Caporali S, Harrington CF, Singh R, Finch D, D’Atri S, Farmer PB, Johnston PG, McKenna WG, Dianov G, Sharma RA. Cells deficient in the base excision repair protein. DNA polymerase beta, are hypersensitive to oxaliplatin chemotherapy, Oncogene. 2010;29:463–468. doi: 10.1038/onc.2009.327. [DOI] [PubMed] [Google Scholar]
- 137.Ochs K, Sobol RW, Wilson SH, Kaina B. Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Research. 1999;59:1544–1551. [PubMed] [Google Scholar]
- 138.Moynihan K, Elion GB, AliOsman F, Marcelli S, Keir S, Bigner DD, Friedman HS. Enhancement of melphalan activity by inhibition of DNA polymerase-alpha and DNA polymerase-beta. Cancer Chemotherapy and Pharmacology. 1996;38:349–354. doi: 10.1007/s002800050494. [DOI] [PubMed] [Google Scholar]
- 139.Raaphorst GP, Ng CE, Yang DP. Comparison of response to radiation, hyperthermia and cisplatin in parental and polymerase beta knockout cells. International Journal of Hyperthermia. 2002;18:33–39. doi: 10.1080/02656730110072352. [DOI] [PubMed] [Google Scholar]
- 140.Horton JK, Baker A, Vande Berg BJ, Sobol RW, Wilson SH. Involvement of DNA polymerase beta in protection against the cytotoxicity of oxidative DNA damage. Dna Repair. 2002;1:317–333. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 141.Horton JK, Srivastava DK, Zmudzka BZ, Wilson SH. Strategic down-regulation of DNA Polymerase beta by antisense RNA sensitizes mammalian cells to specific DNA damaging agents. Nucleic Acids Research. 1995;23:3810–3815. doi: 10.1093/nar/23.19.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boudsocq F, Benaim P, Canitrot Y, Knibiehler M, Ausseil F, Capp JP, Bieth A, Long C, David B, Shevelev I, Frierich-Heinecken E, Hubscher U, Amalric F, Massiot G, Hoffmann JS, Cazaux C. Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase beta. Molecular Pharmacology. 2005;67:1485–1492. doi: 10.1124/mol.104.001776. [DOI] [PubMed] [Google Scholar]
- 143.Daube SS, Tomer G, Livneh Z. Translesion replication by DNA polymerase delta depends on processivity accessory proteins and differs in specificity from DNA polymerase beta. Biochemistry. 2000;39:348–355. doi: 10.1021/bi9917784. [DOI] [PubMed] [Google Scholar]
- 144.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Development. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 145.Sargent JM, Elgie AW, Williamson CJ, Taylor CG. Aphidicolin markedly increases the platinum sensitivity of cells from primary ovarian tumours. British Journal of Cancer. 1996;74:1730–1733. doi: 10.1038/bjc.1996.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Odwyer PJ, Moyer JD, Suffness M, Harrison SD, Cysyk R, Hamilton TC, Plowman J. Antitumor-Activity and Biochemical Effects of Aphidicolin Glycinate (Nsc-303812) Alone and in Combination with Cisplatin In-Vivo. Cancer Research. 1994;54:724–729. [PubMed] [Google Scholar]
- 147.Damia G, Tagliabue G, Zucchetti M, Davoli E, Sessa C, Cavalli F, Dincalci M. Activity of Aphidicolin Glycinate Alone Or in Combination with Cisplatin in A Murine Ovarian Tumor Resistant to Cisplatin. Cancer Chemotherapy and Pharmacology. 1992;30:459–464. doi: 10.1007/BF00685597. [DOI] [PubMed] [Google Scholar]
- 148.Beketicoreskovic L, Osmak M. Modulation of Resistance to Cisplatin by Amphotericin-B and Aphidicolin in Human Larynx-Carcinoma Cells. Cancer Chemotherapy and Pharmacology. 1995;35:327–333. doi: 10.1007/BF00689453. [DOI] [PubMed] [Google Scholar]
- 149.Speit G, Schutz P, Hoffmann H. Enhancement of genotoxic effects in the comet assay with human blood samples by aphidicolin. Toxicology Letters. 2004;153:303–310. doi: 10.1016/j.toxlet.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 150.Zhang NX, Lu XY, Legerski RJ. Partial reconstitution of human interstrand cross-link repair in vitro: characterization of the roles of RPA and PCNA. Biochemical and Biophysical Research Communications. 2003;309:71–78. doi: 10.1016/s0006-291x(03)01535-3. [DOI] [PubMed] [Google Scholar]
- 151.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. Journal of Clinical Oncology. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 152.Shim HJ, Yun JY, Hwang JE, Bae WK, Cho SH, Lee JH, Kim HN, Shin MH, Kweon SS, Lee JH, Kim HJ, Chung IJ. BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Science. 2010;101:1247–1254. doi: 10.1111/j.1349-7006.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Molecular Cancer. 2005;4 doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang R, Niu YJ, Zhou YK. Increase the cisplatin cytotoxicity and cisplatin-induced DNA damage in HepG2 cells by XRCC1 abrogation related mechanisms. Toxicology Letters. 2010;192:108–114. doi: 10.1016/j.toxlet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 155.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutation Research-Dna Repair. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 156.Zhu GY, Lippard SJ. Photoaffinity Labeling Reveals Nuclear Proteins That Uniquely Recognize Cisplatin-DNA Interstrand Cross-Links. Biochemistry. 2009;48:4916–4925. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]