Figure 1.
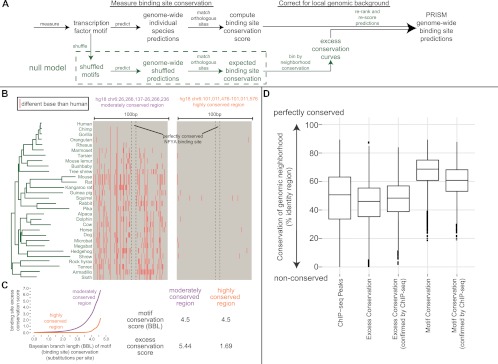
PRISM excess conservation rescoring favors predicted transcription factor binding sites conserved above their local environment. (A) Excess conservation uses the abundance of conserved binding site predictions for shuffled versions of the input motif, in similarly conserved 100-bp genomic neighborhoods, to rescore conserved binding site predictions (framework in green). (B) The NFYA binding site motif is equally conserved in the two shown loci. Yet it is intuitively appealing to consider the left, less conserved 100-bp neighborhood, more likely to conserve an actual NFYA site. (C, left) Excess conservation plots made from all 100-bp neighborhoods conserved like the two loci in panel B. The y-axis is −log10 of the likelihood of a shuffled NFYA motif to achieve the motif conservation score on the x-axis or higher by chance. (Right) Because shuffled versions of NFYA more easily achieve high motif conservation scores in loci like the right locus of panel B, the excess conservation score of this NFYA prediction is lower. (D) Excess conservation rescoring is shown to correct motif conservation-only binding site predictions toward the conservation profile observed in real ChIP-seq peaks. It also outperforms it in area-under-the-curve analysis of 44 (94%) of 47 analyzed ChIP-seq sets (see text).
