Figure 2.
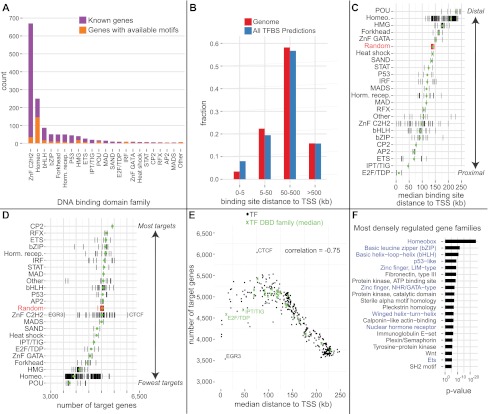
Genome-wide excess conservation binding site predictions reveal fundamental properties of mammalian transcription regulation. (A) The curated library of 332 nonredundant high-quality transcription factor (TF) motifs includes members of all major DNA binding domain (DBD) families. (B) Distributions of all genomic bases (red) and all conserved binding site predictions (blue) as a function of distance from the transcription start site (TSS). While predictions are 2.3-fold enriched in the proximal promoter, >90% of them are distal. (C) Different DNA binding domain families exhibit different binding distance preferences relative to the TSS. (Black ticks) Median distances per motif; (green dot) the family median; random is the median of 332 uniform shuffles. (D) Number of predicted target genes for the different TF DBD families. Black ticks, green dots, and random are as in panel C. POUs and Homeodomains cluster the most around target genes, while CTCF is at the opposite extreme. (E) Distance to TSS and number of target genes have a strong inverse correlation. (F) Transcription factors (blue) are the most densely regulated gene families in the human genome, as measured by the fraction of base pairs in the gene's regulatory domain covered by a binding site prediction. Shown are all nonredundant significant terms after Bonferroni correction (see text).