Figure 3.
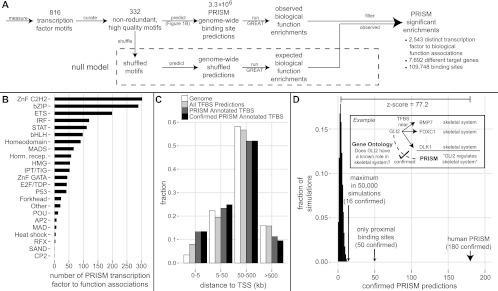
PRISM transcription factor and binding site function predictions. (A) PRISM combines excess conservation binding site prediction (Fig. 1) with GREAT function prediction from proximal and distal sites into a novel statistical framework to arrive at thousands of transcription factor (TF) function predictions, at a false discovery rate of 16%. Numbers are summed over human and mouse (see text). (B) Distribution of PRISM human TF function prediction across the major DNA binding families. (C) Most of the binding sites that support PRISM predictions—including high confidence confirmed predictions—are distal from putative target genes. (D) PRISM predictions are highly enriched for support by previous literature. The GREAT ontologies tag the transcription factor itself with the function predicted by PRISM as enriched among its target genes 180 (11%) times, Z-score = 77, P < 1/50,000 simulation runs (red); 3.6-fold enriched over using only proximal binding sites.
