Abstract
Objective
To review the literature on clinical decision support (CDS) for genetically guided personalized medicine (GPM).
Materials and Methods
MEDLINE and Embase were searched from 1990 to 2011. The manuscripts included were summarized, and notable themes and trends were identified.
Results
Following a screening of 3416 articles, 38 primary research articles were identified. Focal areas of research included family history-driven CDS, cancer management, and pharmacogenomics. Nine randomized controlled trials of CDS interventions for GPM were identified, seven of which reported positive results. The majority of manuscripts were published on or after 2007, with increased recent focus on genotype-driven CDS and the integration of CDS within primary clinical information systems.
Discussion
Substantial research has been conducted to date on the use of CDS to enable GPM. In a previous analysis of CDS intervention trials, the automatic provision of CDS as a part of routine clinical workflow had been identified as being critical for CDS effectiveness. There was some indication that CDS for GPM could potentially be effective without the CDS being provided automatically, but we did not find conclusive evidence to support this hypothesis.
Conclusion
To maximize the clinical benefits arising from ongoing discoveries in genetics and genomics, additional research and development is recommended for identifying how best to leverage CDS to bridge the gap between the promise and realization of GPM.
Keywords: Clinical decision support systems, family health history, genetics, genomics, health information technology, personalized medicine
Background
Genetically guided personalized medicine (GPM) entails the delivery of individually tailored medical care that leverages information about each person's unique genetic characteristics.1 The promise of GPM has expanded as advances in genomics have accelerated over the past several decades. This promise of GPM is that research discoveries will one day lead to medical treatments and therapies that are tailored to the individual characteristics of each patient, including clinical data, genetic test results, patient preference, and family health history (FHx). GPM has the potential to increase the efficacy, quality, and value of healthcare by providing individually optimized prevention, diagnosis, and treatment.2
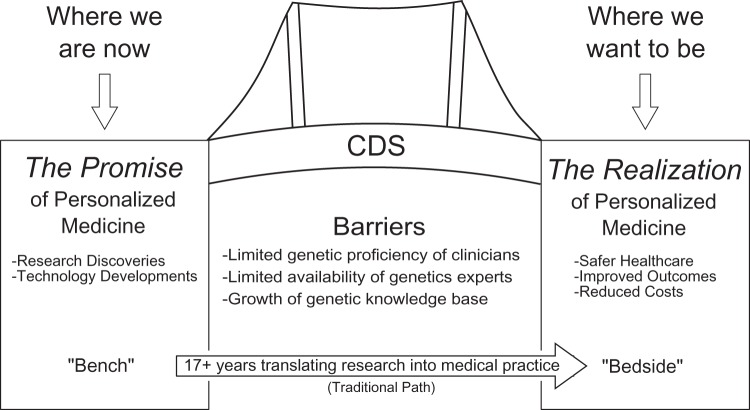
As ongoing research continues to expand the GPM knowledge base, it has become increasingly important to translate this knowledge into routine healthcare practice in order to realize the promise of GPM.3 However, the effective realization of GPM remains very limited.4 While this is partly due to the need for further evidence of the clinical utility and cost effectiveness of a genetically guided approach to patient care, an important additional reason is the need for information systems that assist in the translation of knowledge from bench to bedside.5 Even without the complexity of genetics, it can often take over 15 years to translate research from bench to bedside.6 This translational bottleneck is likely to be an even more significant problem in GPM for the following reasons.
Limited genetic proficiency of clinicians
Many clinicians receive minimal training in clinical genetics. As a result, many physicians lack the confidence and understanding needed for effectively interpreting and using genetic information in their clinical practices.7
Limited availability of genetics experts
Currently, there are about 3000 board-certified genetic counselors8 and approximately 1200 medical geneticists practicing in the USA (S. R. DelBusso, American Board of Medical Genetics Administrator, October 28, 2011, personal communication). The growing utility of genetic information is putting an increasing burden on these professionals. We cannot expect these genetics experts to be readily available each time genetic information should be used to guide medical treatment. For effective, efficient, and widespread clinical use, the burden of genetic interpretation and guidance must be shared by the wider clinical community.
Breadth and growth of genetic knowledge base
There are currently over 2500 clinical genetic tests available to clinicians, encompassing a wide breadth of medical care.9 It is therefore unreasonable to expect a clinician to remember every appropriate genetic test for a particular condition in conjunction with test-specific guidelines for ordering and interpretation. Compounding this issue, the continual growth in the knowledge base and the prospect of full genome sequencing will inevitably overwhelm clinicians' capacities to manage and leverage this information effectively for GPM unless computerized assistance is provided for interpreting and acting on this information.
Various investigators and leaders have identified health information technology as being vital to overcoming these barriers and realizing the promise of GPM.2 10 In particular, clinical decision support (CDS) has been identified as a critical enabler of GPM.11 12 CDS entails providing clinicians, patients, and other healthcare stakeholders with pertinent knowledge and person-specific information, intelligently filtered or presented at appropriate times, to enhance health and healthcare.13 CDS has the capacity to process complex, disparate data and present actionable, standardized, evidence-based recommendations in a way that is usable by a clinician in everyday practice.11 As such, CDS can help bridge the gap between the promise and realization of GPM (figure 1). Given the criticality of CDS for realizing the promise of GPM, and given the lack of a systematic review on this topic, we sought in this paper to assess the history and state of CDS for GPM through a systematic review of the literature.
Figure 1.
Clinical decision support (CDS) as bridge overcoming barriers to genetically guided personalized medicine.
Methods
Data sources and inclusion criteria
We searched MEDLINE and Embase from 1990 to 2011 using a search strategy adapted from previous systematic reviews of CDS,14 genetic health services,15 and FHx16 (see supplementary appendix, available online only, for full search strategy). The final literature search was conducted on June 1, 2012. The inclusion criteria for the review were as follows: English article; human focus; manuscript in peer-reviewed journal; and primary focus on the use of computers to deliver genetically guided, patient-specific assessments and/or recommendations to healthcare providers and/or patients to guide clinical decision-making, as further defined in Box 1.
Box 1. Manuscript inclusion criteria.
-
Definitions:
Healthcare provider = physician, nurse practitioner, physician assistant, registered nurse, or genetic counselor
Genetic factor = genotype, gene expression profile, and/or family health history
-
Universal inclusion criteria:
English article
Human focus
Manuscript in peer-reviewed journal
-
Additional inclusion criteria (at least one):
-
Intervention study evaluating the impact of a CDS system in an actual patient care context
For a comparative intervention study, CDS required to be a part of the primary intervention under evaluation
Excludes laboratory evaluations or simulation studies
Methodology article whose primary focus is on how CDS systems should be designed specifically to support clinical delivery of patient-specific assessments and/or recommendations guided by genetic factors. Includes system description articles.
-
For all identified references, the authors reviewed titles, index terms, and available abstracts to determine if the articles appeared to meet all inclusion criteria. If insufficient information was available to make a confident decision at this stage, the article was included for full-text retrieval. Each full-text article was then reviewed to determine its final inclusion status.
Data abstraction
For each of the articles that met the inclusion criteria listed above, we abstracted data on the clinical application area, CDS type, genetic information used, primary users, article type, study location, CDS purpose, and notable informatics aspects. CDS type was defined as being either stand-alone CDS or integrated CDS. A stand-alone CDS system is a CDS system that exists in isolation from a primary clinical information system containing relevant patient data, such as an electronic health record (EHR) system. A stand-alone CDS system requires manual data input before a CDS result can be produced. In contrast, an integrated CDS system is integrated with a primary clinical information system such as an EHR system or a computerized provider order entry system to aggregate necessary patient-specific information automatically and to provide guidance within routine clinical workflows. Clinical application area was defined as the clinical domain targeted by the CDS intervention. Article type consisted of system description papers and evaluation studies of various types (eg, qualitative evaluation, randomized controlled trial). Genetic information used consisted of FHx, genotype, or both. Primary users were defined as the individuals who primarily entered information and received the results. Study location was the country or region where the research was conducted. CDS purpose identified the role of the CDS system within the context of clinical decision-making. A notable informatics aspect was also abstracted if a manuscript utilized a methodology that was considered to be of potential interest to an informatics audience. For intervention studies, additional details regarding the study size and study outcomes were abstracted.
Data analysis and presentation
Using the abstracted attributes, the manuscripts were grouped into logical categories, primarily according to CDS type and clinical application area. The findings from these manuscripts were summarized through tables and narrative discussion. In addition, notable themes and trends were identified and discussed. A quantitative analysis of CDS trials to identify features predictive of trial outcomes was considered.14 However, due to the limited sample size of CDS trials available, such a quantitative analysis of potential success factors was not feasible.
Results
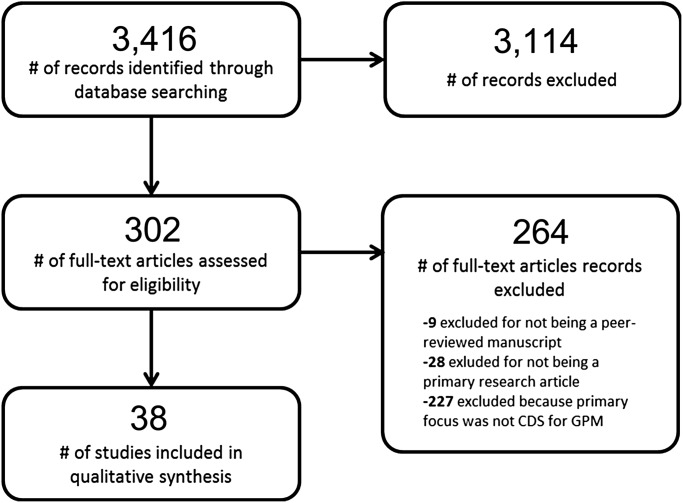
The initial MEDLINE and Embase searches identified 3416 potentially relevant articles. During the title and abstract review, 82 articles were rejected for not being in English, 504 articles were rejected because they were not focused on humans, 34 articles were rejected for not being a peer-reviewed manuscript, and 2494 articles were rejected because the primary focus of the work was not on the use of computers to deliver genetically guided, patient-specific assessments and/or recommendations. The remaining 302 articles underwent full-text review, at which stage 37 articles were rejected for not being a peer-reviewed primary research article and 227 articles were rejected because the primary focus of the work was not on the use of computers to deliver genetically guided, patient-specific care guidance (figure 2). The final set of included manuscripts consisted of 38 primary research articles.17–54 The manuscripts included were published from 1990 to 2011, with the majority of manuscripts published on or after 2007. Provided below is a summary and analysis of these earlier works, grouped primarily by CDS type and area of clinical focus.
Figure 2.
Manuscript selection process. CDS, clinical decision support; GPM, genetically guided personalized medicine.
CDS systems for genetically guided cancer management
Genetically guided cancer management was the focus of 22 primary research articles summarized in tables 1–4.17–37 54 These manuscripts include six manuscripts related to the Risk Assessment in Genetics (RAGs) system for providing FHx-driven CDS (table 1),17–21 54 six manuscripts on other FHx-driven CDS tools for breast cancer management (table 2),22–27 four manuscripts on genotype-driven CDS tools for breast cancer management (table 3),28–31 and six additional manuscripts on GPM CDS tools for non-breast cancer management (table 4).32–37
Table 1.
Summary of primary research on CDS systems for cancer-related GPM: RAGs system for providing FHx-driven CDS
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| Coulson, 200117; RAGs | System description of RAGs, which was designed to help GPs build a family pedigree, calculate genetic risk, and obtain guideline-based care recommendations | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | System description | RAGs uses PROforma, an argumentation-based technology for CDS |
| Emery, 199918; RAGs | A qualitative evaluation of RAGs, which found that the system was easy to use by GPs and served as an appropriate application of information technology to assist with clinical care | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | Qualitative study | RAGs uses PROforma, an argumentation-based technology for CDS |
| Glasspool, 200154; RAGs | System description of RAGs, which uses an argumentation approach to assess genetic risk and provide detailed qualitative explanation with referral advice | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | System description | RAGs uses PROforma, an argumentation-based technology for CDS |
| Emery, 200019; RAGs | Comparative analysis of RAGs, Cyrillic (a commercially available pedigree drawing system), and pen and paper for use by GPs. This study demonstrated that RAGs significantly improved pedigree accuracy and produced more appropriate management decisions than the other two methods. Furthermore, 92% of GPs preferred RAGs to the other methods | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | Comparative study | RAGs uses PROforma, an argumentation-based technology for CDS |
| Emery, 200520; GRAIDS | System description of GRAIDS, a next-generation FHx CDS tool that built on both RAGs and Cyrillic and provided an enhanced user interface for GPs to assess familial cancer risk | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | System description | Server-based application that provides both heuristic and statistical risk assessment |
| Emery, 200721; GRAIDS | A cluster RCT of GRAIDS conducted across 45 GP teams in the UK. GRAIDS significantly increased the number of referrals to the regional genetics clinic (p=0.001), with the referrals being significantly more likely to be consistent with referral guidelines (p=0.006). Moreover, patients referred from GRAIDS practices had significantly lower cancer worry scores at the point of referral (p=0.02) | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for familial breast, ovarian, and colorectal cancer | RCT | Server-based application that provides both heuristic and statistical risk assessment |
CDS, clinical decision support; FHx, family health history; GP, general practitioner; GPM, genetically guided personalized medicine; GRAIDS, Genetic Risk Assessment in an Intranet and Decision Support; RAGs, Risk Assessment in Genetics; RCT, randomized controlled trial.
Table 2.
Summary of primary research on CDS systems for cancer-related GPM: other FHx CDS tools for breast cancer management
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| Tsouskas, 199722 | Evaluation of a CDS tool that used patient-specific breast cancer risk information, including FHx, to identify patients at high risk of breast cancer. The system identified nine out of 10 women with breast cancers in this study | Clinicians in Europe | FHx | No | Assessment of patient risk for breast cancer | Validation study | Expert system developed using a variant of the BASIC programming language |
| Berry, 200223; BRCAPRO | Evaluation of BRCAPRO, which predicted the probability of carrying a BRCA mutation based on a patient's FHx. BRCAPRO was effective in predicting the probability of carrying the BRCA mutation | Clinicians in USA | FHx | No | Assessment of patient risk for breast cancer | Validation study | Probability calculated using Bayesian updating |
| Wilson, 200624 | RCT of a stand-alone breast cancer CDS tool to guide referrals in everyday GP practices. The study consisted of 86 GP practices. The CDS system did not result in a statistically significant improvement, due largely to the limited awareness and adoption of the tool by GPs | GPs in UK | FHx | No | Assessment of patient risk and provision of management recommendations for breast cancer | RCT | The deployment of the CDS system was purposely pragmatic and did not involve extensive workflow integration measures |
| Matloff, 200726 | System description of a tool to provide patient-specific predictions of women's future risks for breast cancer, heart disease, osteoporosis, and endometrial cancer utilizing personal and FHx | Genetic counselors in USA | FHx | No | Assessment of patient risk for breast cancer | System description | System used a Markov model |
| Matloff, 200625 | RCT of a CDS tool used by genetic counselors26 to enable personalized risk assessment and genetic counseling. The trial involved 48 cancer-free, post-menopausal women with a first-degree relative of breast cancer who were contemplating the use of alternative menopausal therapy options. This trial found that patients in the intervention group had increased knowledge and a lower, more accurate perceived risk of developing breast cancer compared to the control group | Genetic counselors in USA | FHx | No | Assessment of patient risk for breast cancer | RCT | System used a Markov model |
| Ozanne, 200927; Hughes Risk Apps | System description of HughesRiskApps and evaluation of its impact at a community hospital. The CDS system significantly increased the number of patients seen for risk consultation and genetic test ordering. The implementation improved efficiency in several ways and did not require significant investment in capital or personnel | Clinicians in USA | FHx | No | Assessment of patient risk and provision of management recommendations for breast cancer | System description; pre–post comparison | Used tablet computers to collect information from patients. Used Health Level 7 compliant information models. |
CDS, clinical decision support; FHx, family health history; GP, general practitioner; GPM, genetically guided personalized medicine; RCT, randomized controlled trial.
Table 3.
Summary of primary research on CDS systems for cancer-related GPM: genotype-driven CDS tools for breast cancer management
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| Schwartz, 200928 | RCT of patient-facing tool that captured patient-specific information and provided tailored content about risks, benefits and management options based on the patients' particular situations. This study found that among 214 BRCA-positive women who were initially undecided about how to manage their breast cancer risk, patients who used the CDS tool were more likely to reach a management decision (p=0.001), had decreased decision conflict (p=0.002), and increased satisfaction (p=0.002) compared to women who did not use the CDS tool | Patients in USA | Genotype | No | Provision of management recommendations for breast cancer | RCT | CD-ROM based, patient-directed decision aid |
| Hooker, 201129 | Longitudinal RCT of patient-facing BRCA decision aid.28 This study showed significantly higher cancer-specific distress (p=0.01) and genetic testing-specific distress (p=0.01) among users of the personalized decision aid after one month. Distress levels between groups were the same after 12 months | Patients in USA | Genotype | No | Provision of management recommendations for breast cancer | RCT | CD-ROM based, patient-directed decision aid |
| Glasspool, 200730; REACT | System description of REACT (Risks, Events, Actions and their Consequences over Time), a breast cancer CDS tool with a graphical timeline display to model real-time changes in lifetime risk as a result of risk-reduction interventions such as tamoxifen therapy, hormone therapy, and mastectomy | Genetic counselors in UK | Genotype | No | Prediction of response to treatment for breast and ovarian cancer | System description | Graphical display of risk changes dynamically based on selected interventions |
| Glasspool, 201031; REACT | Qualitative study of REACT by eight genetic counselors.30 Most counselors found REACT effective for genetic risk management, although there were concerns related to the tool's potential to alter the dynamics of the clinician–patient interaction | Genetic counselors in UK | Genotype | No | Prediction of response to treatment for breast and ovarian cancer | Qualitative study | Graphical display of risk changes dynamically based on selected interventions |
CDS, clinical decision support; GPM, genetically guided personalized medicine; RCT, randomized controlled trial; REACT, Risks, Events, Actions and their Consequences over Time.
Table 4.
Summary of primary research on CDS systems for cancer-related GPM: CDS for other cancers
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| Evans, 199532 | Description of a FHx CDS system developed for a hereditary cancer consulting service. The system collected FHx information, evaluated the FHx for familial risk patterns, and produced preliminary risk assessment and management recommendations. The system resulted in a significant reduction in time spent on cases | Hereditary cancer consulting service in USA | FHx | No | Assessment of patient risk and provision of management recommendations for hereditary cancer | System description; impact observation | Expert rule-based system that modeled the pattern recognition capabilities of clinical geneticists |
| Bianchi, 200733; CRCAPRO | Evaluation of CRCAPRO, which used FHx of colorectal and endometrial cancers to identify patients with Lynch syndrome. This study showed that CRCAPRO has low sensitivity and specificity | Clinicians in UK | FHx | No | Assessment of patient risk for colorectal cancer | System validation | Probability calculated using Bayesian updating |
| Overbeek, 201034 | RCT of electronic reminders to pathologists to consider Lynch syndrome genetic testing among newly diagnosed colon cancer patients based on FHx. The CDS reminder intervention in 12 pathology laboratories significantly improved pathologists' recognition of patients at risk for Lynch syndrome (OR 2.8; 95% CI 1.1 to 7.0) and increased use of genetic testing (OR 4.1; 95% CI 1.3 to 13.2) | Pathologists in Europe | FHx | Yes | Provision of management recommendations for colorectal cancer | RCT | Electronic reminders provided through health information system |
| Picone, 201135; NeoMark | System description of NeoMark, a web-based tool that combined medical images, genetic markers, and other patient data before and after treatment of oral cavity squamous cell carcinoma to predict reoccurrence | Clinicians in Europe | Genotype | No | Assessment of patient risk for oral cancer | System description | Uses a service-oriented, modular architecture |
| Hendershot, 201036 | RCT of a web-based genetic feedback intervention involving 200 college students of Asian descent. The system provided personalized alcohol-related health risk information and feedback based on the patient's genotype. The tool resulted in significant reductions in drinking (p=0.02) among participants with the genotype associated with higher risk of alcohol-related cancer | Patients in USA | Genotype | No | Assessment of patient risk; reduction of risky behavior (alcohol consumption) for alcohol-related cancer | RCT | Web-based intervention |
| Wakefield, 201137 | System description and pilot usability test of an online CDS tool that presented 22 men with age and family history-specific prostate cancer risk information and management recommendations. Most participants preferred this method for receiving prostate cancer information | Patients in Australia/New Zealand | FHx | No | Assessment of patient risk and provision of management recommendations for prostate cancer | System description; pilot usability test | Online decision aid using a Markov model |
CDS, clinical decision support; FHx, family health history; GPM, genetically guided personalized medicine; RCT, randomized controlled trial.
RAGs system for providing FHx-driven CDS
Some of the earliest and most comprehensive research on the use of CDS to support GPM was conducted by Emery55 (table 1), who identified that existing systems were not designed for primary care and that none provided patient management advice based on calculated risk. To address this gap, Emery developed a system known as RAGs, which helped general practitioners (GPs) in the UK collect FHx relevant to familial breast, ovarian, and colorectal cancer and provided appropriate management guidance, primarily regarding guideline-based specialist referrals.17–19 54 A later extension of the RAGs system was referred to as the GRAIDS system.20 21 This body of work included several favorable evaluations of these systems,18 19 21 including a cluster randomized controlled trial (RCT) across 45 GP teams that found that GRAIDS significantly increased the proportion of patients referred appropriately to the regional genetics clinic according to evidence-based practice guidelines.21
Other FHx CDS tools for breast cancer management
Beyond the work of Emery,55 CDS research for GPM has focused heavily on breast cancer management (table 2). Risk assessment tools for breast cancer can enable personalized care according to an individual's level of risk.22 23 An RCT conducted in the UK found that a stand-alone breast cancer CDS tool had limited impact due to lack of awareness and use by GPs.24 At the same time, a stand-alone CDS tool that calculated risks for breast cancer, heart disease, osteoporosis, and endometrial cancer was shown in an RCT to enhance the effectiveness of genetic counselors using the system.25 26 Another stand-alone CDS system that has been found to be beneficial is HughesRiskApps, which collects relevant FHx information and provides clinicians with various tools to support the management of patients. An observational implementation study of this tool in a community hospital setting found significant adoption and impact.27
Genotype-driven CDS tools for breast cancer management
Several investigators have developed CDS systems that support treatment and decision-making once mutations have been identified in the breast cancer (BRCA) genes (table 3). In the UK, Glasspool and colleagues30 31 developed a CDS tool known as REACT (Risks, Events, Actions and their Consequences over Time), which used a graphical timeline display to model real-time changes in lifetime risks as a result of risk-reduction interventions for breast cancer and ovarian cancer. In addition, several patient-directed, stand-alone CDS systems have been developed for improving risk communication and decision-making in breast cancer management based on BRCA genotype.28 29
CDS for other cancers
Besides breast cancer, other cancers have been the focus of CDS research and development (table 4). Most of this CDS research for other cancers has involved colorectal cancer, and in particular Lynch syndrome—a strongly heritable type of colorectal cancer.32–34 Of note, the RAGs and GRAIDS systems described earlier supported both breast cancer and colorectal cancer management.17–21 54 An additional CDS system investigated for colorectal cancer management is CRCAPRO, similar to BRCAPRO, which used FHx to identify patients at risk of hereditary colorectal cancer.33 In addition, a group in the Netherlands developed a CDS intervention to remind pathologists to order Lynch syndrome genetic testing among patients who met certain criteria, one of which was a suspicious FHx. This intervention significantly improved pathologists' recognition of patients at risk of Lynch syndrome.34 Moreover, Dr Henry Lynch, for whom Lynch syndrome is named, developed a CDS system for supporting his hereditary cancer consulting service. This CDS system expedited clinicians' decision-making processes and resulted in a significant reduction in time spent on cases.32
Similar to the stand-alone CDS systems for breast cancer management described earlier,28 29 stand-alone CDS tools have been shown to be useful for the management of other types of cancers, including prostate cancer37 and alcohol-related cancers.36 These studies included an RCT that showed that a patient-directed, genotype-driven CDS tool for alcohol-related cancer risk significantly reduced alcohol consumption by patients at increased genetic risk.36 These studies, as well as the previous studies on breast cancer,28 29 showed that patient-directed CDS systems can be clinically useful.
CDS for pharmacogenomics
Pharmacogenomics, the practice of tailoring drug therapy to the patient's unique genetic characteristics, can be a complicated process; genetically guided CDS offers a solution for simplifying this process. Table 5 summarizes the six primary research articles identified on this topic.38–43 These studies include a description and validation of a CDS system for genetically guided treatment of HIV infections,38 as well as an RCT that found that genotyping combined with CDS-guided therapy improved outcomes over standard of care.39 Outside of HIV therapy, other investigators focused on how CDS for pharmacogenomics could be integrated with primary clinical information systems such as computerized provider order entry systems.40 42 43 These studies evaluated considerations such as developing the underlying pharmacogenomics knowledge base,40 representation of genetic information in the EHR for supporting pharmacogenomics CDS,42 and the availability of patient data required for pharmacogenomics within the EHR.43 The lone stand-alone system for pharmacogenomics used genotype and clinical data to estimate and graphically represent a patient's plasma warfarin concentration over time.41
Table 5.
Summary of primary research on CDS systems for pharmacogenomics
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| Pazzani, 199738; CTSHIV | System description of the CTSHIV CDS program which manages HIV genome data and makes virus-specific therapeutic recommendations | Clinicians in USA | HIV genotype | No | Provision of management recommendations for HIV | System description | Uses a backward chaining expert system |
| Tural, 200239; RetroGram | RCT of genotyping accompanied by RetroGram, which ranked drug suitability based on the HIV genotype. This study showed that genotyping combined with RetroGram use improved HIV therapy outcomes over standard of care (p<0.05) | Clinicians in Europe | HIV genotype | No | Provision of management recommendations for HIV | RCT | Contains approximately 200 rules based on the scientific literature |
| Swen, 200840 | Description of how the Royal Dutch Association for the Advancement of Pharmacy developed guidelines for the use of genetic information for drug prescribing and integrated these guidelines into automated drug prescription and medical surveillance systems for nationwide use | Clinicians and pharmacists in Europe | Genotype | Yes | Alert on gene-drug interactions for pharmacogenomics | System description | Recommendations incorporated into the G-standard, an electronic drug database used for CDS |
| Bon Homme, 200841 | System description of prototype CDS tool for personalized warfarin therapy that combined genetic and clinical data to estimate the required warfarin dose and the patient's plasma warfarin concentration | Clinicians in USA | Genotype | No | Therapeutic dose guidance for warfarin | System description | Provides a graphical display of estimated plasma warfarin concentration over time |
| Deshmukh, 200942 | This study compared the use of a single nucleotide polymorphism data model to the use of an allele data model for CDS computation in an EHR system. While there were statistically significant differences in computation time, this did not translate into significant differences in the overall clinician ordering time | Clinicians and pharmacists in USA | Genotype | Yes | Alert on gene–drug interactions for pharmacogenomics | Comparative study on genotype data representation | CDS rules developed within the Cerner EHR environment |
| Overby, 201043 | This study found that the Pharmacogenomics Knowledge Base was a good source for pharmacogenomics knowledge and that sufficient clinical data existed in the local EHR system to support 50% of the pharmacogenomic knowledge in drug labels that are capable of being expressed as CDS rules | Clinicians in USA | Genotype | Yes | Provision of therapy guidance for pharmacogenomics | Feasibility study | The MINDscape EHR system was used in the study |
CDS, clinical decision support; CTSHIV, Customized Treatment Strategies for HIV; EHR, electronic health record; RCT, randomized controlled trial.
Other CDS systems for GPM
Table 6 summarizes the 10 primary research articles that were neither cancer specific nor focused on pharmacogenomics.44–53 As with CDS for cancer, there has been a substantial focus on FHx-driven CDS for other medical conditions. For example, a tool called GenInfer considered FHx and calculated inheritance risks for genetic diseases,44 and FHx-driven CDS was included as a part of the National Russian Genetic Register.45 46 Beyond these system descriptions, recent studies of FHx-driven CDS have focused on impact evaluation, with mixed results.48 49 51
Table 6.
Summary of primary research on GPM CDS systems for other conditions
| Citation and name of system (if applicable) | Manuscript summary and trial details (if applicable) | Users and study location | Genetic information used | Integrated with primary clinical information system | CDS purpose and clinical focus | Manuscript type | Notable informatics aspect |
| FHx-driven CDS systems | |||||||
| Harris, 199044; GenInfer | System description of the GenInfer program, which used FHx information along with other inheritance factors to calculate genetic risks and probabilities of inheritance | Clinicians in USA | FHx | No | Assessment of patient risk for inherited disease | System description | Based on Pearl's algorithm for fusion and propagation in a probabilistic belief network |
| Kobrinskii, 199745 and Kobrinsky, 199846; National Russian Genetic Register | Description of the information system used by Russia's federal genetics center to manage patients across Russia in need of genetics care. This system supported pedigree creation, cytogenetic analysis, risk assessment, and information support | Genetics specialists in Europe | FHx | No | Assessment of patient risk for inherited disease | System description | Utilized both server–client and local deployment models |
| Orlando, 201148; MeTree | System description of MeTree, a tool that evaluates FHx and provides management recommendations regarding various heritable conditions for patients and clinicians. Also provides the protocol for a planned evaluation of the tool in North Carolina primary care clinics | Patients and clinicians in USA | FHx | No | Assessment of patient risk and provision of management recommendations for inherited disease | System description; evaluation protocol description | Patient-driven application that provides CDS as a printout |
| Rubinstein, 201149; CDC Family Healthware | RCT with 3284 participants of the CDC Family Healthware tool, which provides personalized screening recommendations for multiple heritable conditions based on FHx. Both intervention and control groups showed improved adherence to screening recommendations compared to the baseline time period, but there was no significant difference between the intervention and control groups | Patients in USA | FHx | No | Assessment of patient risk and provision of management recommendations for inherited disease | RCT | A patient-directed, web-based tool |
| Wells, 200751; PREDICT CVD-5 | System description of a real-time CDS system that pulled clinical data from the EHR to calculate cardiovascular disease risk and provide risk management recommendations. A retrospective analysis found that including the patients' ethnicity and FHx into the risk assessment process substantially increased the number of patients eligible for drug treatment and lifestyle management | Clinician in Australia and New Zealand | FHx | Yes | Assessment of patient risk for heart disease | System description; retrospective analysis | Integrated with the MedTech practice management system |
| Genotype-driven CDS systems | |||||||
| Iavindrasana, 200852; @neurIST | System description of @neurIST, a CDS system which collects genetic data, radiological data, and clinical data from clinical information systems to provide CDS regarding intracranial aneurisms | Clinicians in Europe | Genotype | Yes | Provision of management recommendations for intracranial aneurism | System description | Uses a service- oriented, standards-based approach |
| Kalatzis, 200947 | System description of a point-of-care portable medical device that integrates clinical data with genetic data obtained from a miniature diagnostic system to produce a diagnosis for rheumatoid arthritis and multiple sclerosis | Clinicians in Europe | Genotype | No | Diagnostic assistance for arthritis and multiple sclerosis | System description | A combination of artificial neural networks, decision trees, and support vector machines was found to have the best performance |
| Scheuner, 200953 | A survey of health professionals, genetics experts, and EHR developers regarding the ability of EHR systems to document, organize, and use FHx and genetic information | Clinicians in USA | FHx; genotype | Yes | Assessment of patient risk and provision of management recommendations for genetically-guided personalized medicine | Survey | EHRs were generally perceived as lacking the ability to support genomic medicine |
| Aronson, 201150; GeneInsight | System description of GeneInsight, a platform that provides patient-specific genetic testing reports as well as notifications when the presumed clinical significance of genetic variants change for patients who have been previously tested | Geneticists and other clinicians in USA | Genotype | No | Provision of patient-specific genetic testing reports; notification of changes in clinical significance of genetic variants | System description | Is registered with the Food and Drug Administration as a class I exempt medical device |
CDC, Centers for Disease Control and Prevention; CDS, clinical decision support; EHR, electronic health record; FHx, family health history; GPM, genetically guided personalized medicine; RCT, randomized controlled trial.
Finally, there were four primary research studies on genotype-driven CDS systems not focused on pharmacogenomics or cancer.47 50 52 53 These systems included a CDS system that retrieved genetic, radiological and clinical data from clinical information systems to provide guidance on intracranial aneurism management,52 as well as a portable medical device that integrated clinical and genetic data to provide a diagnosis for rheumatoid arthritis and multiple sclerosis.47 In addition, GeneInsight provides geneticists and other clinicians with patient-specific genetic testing reports, as well as notifications regarding updates to the presumed clinical significance of patients' previously identified genotype.50 Finally, in a survey study, Scheuner and colleagues53 found that clinicians felt their EHR systems could do much more to meet their needs related to GPM.
Trend analysis
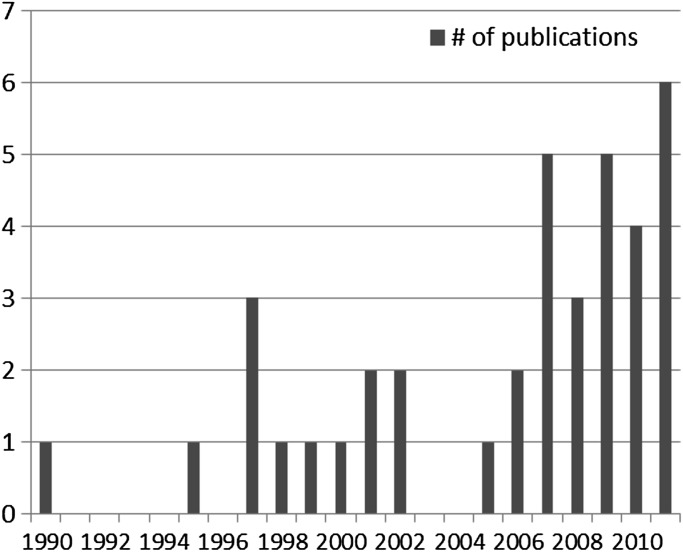
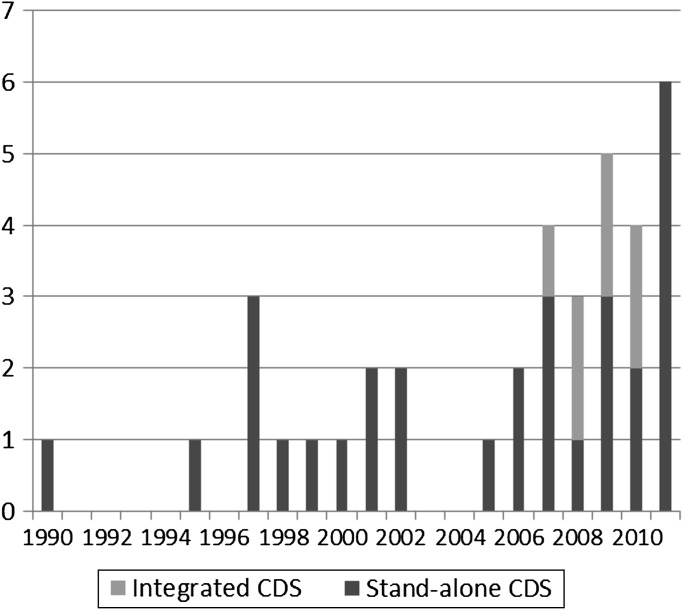
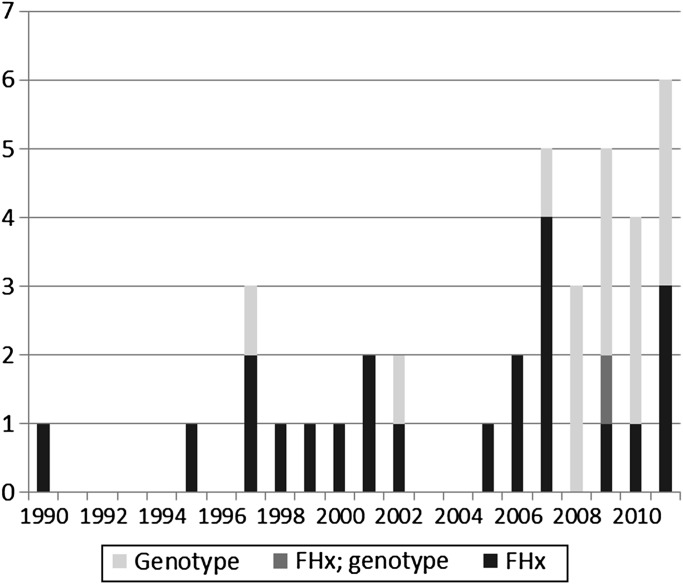
Publication volume on CDS for GPM generally increased over time, with a majority published since 2007 (figure 3). While all publications before 2007 focused on stand-alone CDS, 32% of articles since 2007 focused on integrated CDS (figure 4). Likewise, while 13% of manuscripts before 2007 involved the use of genotype for CDS, 61% of manuscripts since 2007 have involved the use of genotype (figure 5). As noted earlier, a major focus of the literature in this domain has been on FHx CDS, pharmacogenomics, and CDS for cancer management.
Figure 3.
Publications included per year.
Figure 4.
Publications focused on stand-alone versus integrated clinical decision support (CDS).
Figure 5.
Publications focused on family health history (FHx)-driven versus genotype-driven clinical decision support.
Discussion
Summary of findings
In order to learn from past research efforts and to guide future research into the use of CDS to enable GPM, we conducted a systematic review of the literature. Through a literature search spanning from 1990 to 2011, we screened 3416 manuscripts and included 38 primary research articles. A majority of these manuscripts was published from 2007 to 2011, with an increasing shift in focus from FHx CDS to genotype-driven CDS, and from stand-alone CDS to integrated CDS. There have been nine RCTs of CDS interventions for GPM, but most CDS interventions for GPM have not yet been rigorously assessed for their clinical impact.
Strengths and limitations
As one important strength of this study, as far as we are aware, this work represents the first systematic review on CDS for GPM. As such, it contributes an important perspective on a topic that has the potential to have significant impacts in both clinical medicine and biomedical informatics. As a second strength, this systematic review was based on search strategies refined through previous systematic reviews on related topics.14–16 Third, we searched Embase in addition to MEDLINE, so as to provide greater coverage of the international literature. Finally, in addition to providing a summary of relevant manuscripts, this review provides insights and trend analyses that show how this scientific field has developed over time and where the field appears to be headed moving forward.
In terms of limitations, this study does not provide a quantitative meta-analysis of the impact of CDS interventions for GPM. However, such a meta-analysis was not possible due to the limited number of outcome studies in this field and the heterogeneous nature of the various interventions and clinical domains. Second, we only included manuscripts written in English, which may have led to some relevant manuscripts being excluded that were written in a different language. Third, some relevant 2011 articles may not have been indexed by the time of our literature search and therefore erroneously excluded. However, a literature search update in June 2012 added less than 1% to the number of articles we had previously retrieved through March 2012, which suggests that this risk is low. Finally, there is a potential for publication bias with regard to the clinical trials included, in which studies with successful outcomes were more likely to be published than studies with unsuccessful outcomes. There was a potential indication of such a bias, in that seven of nine RCTs evaluated (77%) reported positive results, whereas the expected rate of positive results would more typically be in the range of approximately 60%.56 However, given the limited sample size, the observed discrepancy may simply be due to chance. Moreover, as discussed next, the high rate of successful interventions may be partly explained by the fact that use of many of these systems was required by the study protocol, which improved the systems' likelihood of use and impact.
Consistency of trial findings with expected outcomes
In a previous systematic review of CDS RCTs, we identified the automatic provision of CDS as a part of routine clinical workflow to be a critical predictor of the success or failure of CDS interventions (adjusted OR of 112.1, p<0.00001).14 While automatic provision of CDS was not a guarantee of success in this systematic review, a lack of this feature was associated with negative outcomes in all cases, generally due to the lack of use of the system.14 Moreover, a later RCT specifically evaluating the importance of automatic provision of CDS directly confirmed this finding.57
On initial examination, the results of the present systematic review seemed to contradict this finding, as we found several RCTs in which stand-alone CDS interventions for GPM were not provided automatically as a part of routine clinical workflow but resulted in positive improvements in clinical practice.21 25 28 29 36 39 However, in all but one of these RCTs,21 use of the CDS system was mandated by the study protocol, which was an exclusion criterion in the previous systematic review that identified the critical importance of the automatic provision of CDS.14 Therefore, we believe it is premature to draw the conclusion that automatic provision is not important when providing CDS for GPM, as it is possible that the same CDS interventions that led to positive results in the studies included would not have led to positive results if use of the system was not mandated by the study protocol, due to lack of awareness and use of the tool. With regard to other, less critical success factors identified in the previous systematic review of CDS interventions,14 we did not find any trends that contradicted those findings. However, the sample size of available CDS trials was too small in this study to allow for any meaningful analysis of these other factors.
Of note, in the RCT of the GRAIDS system for FHx-based CDS, the system did have a positive impact, even though its use was not mandated by the study protocol and the system was not automatically provided as a part of routine clinical workflow.14 However, the use and impact of this system may have been the result of exceptional circumstances specific to the study context and unlikely to be available in a routine clinical practice setting. In particular, in the RCT of the GRAIDS system, designated clinicians were recruited at each practice, received extensive training on GRAIDS, and managed all patients in the practice expressing concern regarding their breast or colorectal cancer FHx.21 This type of resource-intensive deployment strategy may not be feasible outside the context of a research study, as demonstrated in another RCT of a stand-alone breast cancer CDS tool, which had limited impact due largely to the lack of awareness and adoption of the tool by clinicians.24 Therefore, while more evidence is needed before a solid conclusion can be drawn, we found no conclusive evidence that CDS for GPM is unique in terms of the intervention features required for successful outcomes.
Assessment of current research state and required research
In recent years, CDS has been proposed as a promising approach to realizing the promise of GPM.10–12 58–65 However, we identified only 38 primary research articles published from 1990 to 2011 on the design, implementation, use, and evaluation of CDS systems to support genetically guided patient care, which amounts to approximately 1.7 articles per year. Even in the year with the most publications on this topic (2011), we identified only six primary research articles. In particular, we identified only nine RCTs of the impact of CDS systems for GPM, seven articles focused on CDS integrated with primary clinical information systems, and 16 articles involving the use of genotype to drive CDS. Furthermore, few groups have demonstrated how genotype-driven CDS can be integrated into clinical settings and clinical information systems in a scalable, standards-based, and effective manner.40 43 52 53
Given the tremendous volume of research being conducted in the discovery of novel personalized medicine diagnostics and therapeutics, we feel that much more research is required on how CDS can and should be leveraged to take these discoveries and to implement them in routine clinical practice. For example, even for FHx-driven CDS, which is perhaps the most well-established area of research with regard to CDS for GPM, there has been limited research on the optimal use of FHx-driven CDS tools beyond hereditary cancer management. Indeed, given the limited literature available on any one topic, we feel it would be premature to consider any aspect of CDS for GPM to be fully mature and not in need of any further research.
In looking forward, we believe that the largest looming research challenge in terms of CDS for GPM will be the development of effective approaches to manage and utilize whole genome sequence data in the clinical setting. The pursuit of low-cost whole genome sequencing has been a priority research area for many years, such that sequencing costs may be reduced to a level amenable to routine clinical use in the near future.66 While sequencing technologies continue to advance, the informatics capabilities to apply whole genome sequencing data to clinical practice is still in its infancy.67 Indeed, in our systematic review, we did not find a single primary research article addressing this topic. Therefore, we recommend the prioritization and resourcing of this area of research by the scientific community. In particular, to realize the full clinical potential of whole genome sequence data, we believe that approaches will need to be developed for providing advanced CDS capabilities that are integrated with clinical information systems and provided automatically as a part of routine clinical workflow.
Conclusion
The promise of GPM is growing with the recent advances and discoveries in genomics research. With this growth also comes the growing need for translating such discoveries into everyday clinical care, so that we are able to realize the promises of GPM. CDS has the potential to bridge this gap between the promise and realization of GPM. By systematically reviewing the literature in this field and by identifying gaps in required research, we speculate that this paper will assist with efforts to leverage CDS to enable GPM at scale.
Supplementary Material
Footnotes
Contributors: BMW and KK both contributed to the design and conduct of the study, as well as the preparation of the manuscript.
Funding: This study was funded by grant K01HG004645 from the US National Human Genome Research Institute, the University of Utah Department of Biomedical Informatics, and the University of Utah Program in Personalized Health Care. The funding sources played no role in the study design, in the collection, analysis and interpretation of data, in the writing of the manuscript, or in the decision to submit the manuscript for publication. The views expressed in this manuscript are those of the authors alone and do not necessarily reflect the official views of the National Human Genome Research Institute or of the University of Utah.
Competing interests: BMW is the founder and owner of SGgenomics, Inc., which developed ItRunsInMyFamily.com, a patient-centered FHx tool. KK is serving as a consultant to Inflexxion on a project funded by the National Institute on Drug Abuse to develop CDS capabilities for mental healthcare. KK receives royalties for a Duke University-owned CDS technology for infectious disease management known as CustomID that he helped develop. KK was formerly a consultant for Religent, Inc. and a co-owner and consultant for Clinica Software, Inc., both of which provide commercial CDS services, including through use of a CDS technology known as SEBASTIAN that KK developed. KK no longer has a financial relationship with either Religent or Clinica Software.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Individuals interested in the raw data and analyses may contact the corresponding author to obtain such data.
References
- 1. U.S. Department of Health and Human Services Glossary Of Terms For Personalized Health Care Website. http://www.hhs.gov/myhealthcare/glossary/glossary.html (accessed Aug 2012). [Google Scholar]
- 2. Abrahams E, Ginsburg GS, Silver M. The personalized medicine coalition: goals and strategies. Am J Pharmacogenomics 2005;5:345–55 [DOI] [PubMed] [Google Scholar]
- 3. Khoury MJ, McBride CM, Schully SD, et al. The scientific foundation for personal genomics: recommendations from a National Institutes of Health–Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med 2009;11:559–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Offit K. Personalized medicine: new genomics, old lessons. Hum Genet 2011;130:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernald GH, Capriotti E, Daneshjou R, et al. Bioinformatics challenges for personalized medicine. Bioinformatics 2011;27:1741–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balas EA, Boren SA. Managing clinical knowledge for health care improvement. Yearbook of Medical Informatics 2000: Patient-Centered System. Stuttgart, Germany: Schattauer, 2000:65–70 [PubMed] [Google Scholar]
- 7. Rosas-Blum E, Shirsat P, Leiner M. Communicating genetic information: a difficult challenge for future pediatricians. BMC Med Educ 2007;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. American Board of Genetic Counseling, Inc About ABGC. http://www.abgc.net/About_ABGC/GeneticCounselors.asp (accessed Aug 2012).
- 9. GeneTests: Growth of Laboratory Directory. Seattle: University of Washington; http://www.ncbi.nlm.nih.gov/projects/GeneTests/static/whatsnew/labdirgrowth.shtml (accessed Aug 2012). [Google Scholar]
- 10. Glaser J, Henley DE, Downing G, et al. Personalized Health Care Workgroup of the American Health Information Community. Advancing personalized health care through health information technology: an update from the American Health Information Community's Personalized Health Care Workgroup. J Am Med Inform Assoc 2008;15:391–6 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC130085/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawamoto K, Lobach DF, Willard HF, et al. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med Inf Decis Mak 2009;9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Downing GJ, Boyle SN, Brinner KM, et al. Information management to enable personalized medicine: stakeholder roles in building clinical decision support. BMC Med Inf Decis Mak 2009;9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osheroff JA, Teich JM, Middleton B, et al. A roadmap for national action on clinical decision support. J Am Med Inform Assoc 2007;14:141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawamoto K, Houlihan CA, Balas EA, et al. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 2005;330:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: a systematic review. JAMA 2008;299:1320–34 [DOI] [PubMed] [Google Scholar]
- 16. Wilson BJ, Qureshi N, Santaguida P, et al. Systematic review: family history in risk assessment for common diseases. Ann Intern Med 2009;151:878–85 [DOI] [PubMed] [Google Scholar]
- 17. Coulson AS, Glasspool DW, Fox J, et al. RAGs: a novel approach to computerized genetic risk assessment and decision support from pedigrees. Methods Inf Med 2001;40:315–22 [PubMed] [Google Scholar]
- 18. Emery J, Walton R, Coulson A, et al. Computer support for recording and interpreting family histories of breast and ovarian cancer in primary care (RAGs): qualitative evaluation with simulated patients. BMJ 1999;319:32–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emery J, Walton R, Murphy M, et al. Computer support for interpreting family histories of breast and ovarian cancer in primary care: comparative study with simulated cases. BMJ 2000;321:28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emery J. The GRAIDS Trial: the development and evaluation of computer decision support for cancer genetic risk assessment in primary care. Ann Hum Biol 2005;32:218–27 [DOI] [PubMed] [Google Scholar]
- 21. Emery J, Morris H, Goodchild R, et al. The GRAIDS Trial: a cluster randomised controlled trial of computer decision support for the management of familial cancer risk in primary care. Br J Cancer 2007;97:486–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsouskas LI, Liaros A, Tzitzikas J, et al. The use of an expert system of composite risk factors in breast cancer screening. Stud Health Technol Inform 1997;43:859–63 [PubMed] [Google Scholar]
- 23. Berry DA, Iversen ES, Jr, Gudbjartsson DF, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol 2002;20:2701–12 [DOI] [PubMed] [Google Scholar]
- 24. Wilson BJ, Torrance N, Mollison J, et al. Cluster randomized trial of a multifaceted primary care decision-support intervention for inherited breast cancer risk. Fam Pract 2006;23:537–44 [DOI] [PubMed] [Google Scholar]
- 25. Matloff ET, Moyer A, Shannon KM, et al. Healthy women with a family history of breast cancer: impact of a tailored genetic counseling intervention on risk perception, knowledge, and menopausal therapy decision making. J Womens Health 2006;15:843–56 [DOI] [PubMed] [Google Scholar]
- 26. Matloff ET, Shannon KM, Moyer A, et al. Should menopausal women at increased risk for breast cancer use tamoxifen, raloxifene, or hormone therapy? A framework for personalized risk assessment and counseling. J Cancer Educ 2007;22:10–14 [DOI] [PubMed] [Google Scholar]
- 27. Ozanne EM, Loberg A, Hughes S, et al. Identification and management of women at high risk for hereditary breast/ovarian cancer syndrome. Breast J 2009;15:155–62 [DOI] [PubMed] [Google Scholar]
- 28. Schwartz MD, Valdimarsdottir HB, DeMarco TA, et al. Randomized trial of a decision aid for BRCA1/BRCA2 mutation carriers: impact on measures of decision making and satisfaction. Health Psychol 2009;28:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hooker GW, Leventhal KG, DeMarco T, et al. Longitudinal changes in patient distress following interactive decision aid use among BRCA1/2 carriers: a randomized trial. Med Decis Making 2011;31:412–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glasspool DW, Oettinger A, Smith-Spark JH, et al. Supporting medical planning by mitigating cognitive load. Methods Inf Med 2007;46:636–40 [DOI] [PubMed] [Google Scholar]
- 31. Glasspool DW, Oettinger A, Braithwaite D, et al. Interactive decision support for risk management: a qualitative evaluation in cancer genetic counselling sessions. J Cancer Educ 2010;25:312–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans S, Lynch HT, Fusaro RM. Clinical results using informatics to evaluate hereditary cancer risk. Proc Annu Symp Comput Appl Med Care 1995:834–8 [PMC free article] [PubMed] [Google Scholar]
- 33. Bianchi F, Galizia E, Bracci R, et al. Effectiveness of the CRCAPRO program in identifying patients suspected for HNPCC. Clin Genet 2007;71:158–64 [DOI] [PubMed] [Google Scholar]
- 34. Overbeek LI, Hermens RP, van Krieken JH, et al. Electronic reminders for pathologists promote recognition of patients at risk for Lynch syndrome: cluster-randomised controlled trial. Virchows Arch 2010;456:653–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Picone M, Steger S, Exarchos K, et al. Enabling heterogeneous data integration and biomedical event prediction through ICT: the test case of cancer reoccurrence. Adv Exp Med Biol 2011;696:367–75 [DOI] [PubMed] [Google Scholar]
- 36. Hendershot CS, Otto JM, Collins SE, et al. Evaluation of a brief web-based genetic feedback intervention for reducing alcohol-related health risks associated with ALDH2. Ann Behav Med 2010;40:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wakefield CE, Watts KJ, Meiser B, et al. Development and pilot testing of an online screening decision aid for men with a family history of prostate cancer. Patient Educ Couns 2011;83:64–72 [DOI] [PubMed] [Google Scholar]
- 38. Pazzani MJ, See D, Schroeder E, et al. Application of an expert system in the management of HIV-infected patients. J Acquir Immune Defic Syndr Hum Retrovirol 1997;15:356–62 [DOI] [PubMed] [Google Scholar]
- 39. Tural C, Ruiz L, Holtzer C, et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS 2002;16:209–18 [DOI] [PubMed] [Google Scholar]
- 40. Swen JJ, Wilting I, de Goede AL, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther 2008;83:781–7 [DOI] [PubMed] [Google Scholar]
- 41. Bon Homme M, Reynolds KK, Valdes R, Jr, et al. Dynamic pharmacogenetic models in anticoagulation therapy. Clin Lab Med 2008;28:539–52 [DOI] [PubMed] [Google Scholar]
- 42. Deshmukh VG, Hoffman MA, Arnoldi C, et al. Efficiency of CYP2C9 genetic test representation for automated pharmacogenetic decision support. Methods Inf Med 2009;48:282–90 [DOI] [PubMed] [Google Scholar]
- 43. Overby CL, Tarczy-Hornoch P, Hoath JI, et al. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11(Suppl. 9):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harris NL. Probabilistic belief networks for genetic counseling. Comput Methods Programs Biomed 1990;32:37–44 [DOI] [PubMed] [Google Scholar]
- 45. Kobrinskii BA, Tester IB, Demikova NS, et al. A concept of the federal computer system for families with genetic diseases and its practical implementation. Biomed Eng 1997;31:172–4 [Google Scholar]
- 46. Kobrinsky B, Tester I, Demikova N, et al. A multifunctional system of the national genetic register. Stud Health Technol Inform 1998;52:121–5 [PubMed] [Google Scholar]
- 47. Kalatzis FG, Giannakeas N, Exarchos TP, et al. Developing a genomic-based point-of-care diagnostic system for rheumatoid arthritis and multiple sclerosis. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc; 2–6 September 2009, Minneapolis, Minnesota, 2009:827–30 [DOI] [PubMed] [Google Scholar]
- 48. Orlando LA, Hauser ER, Christianson C, et al. Protocol for implementation of family health history collection and decision support into primary care using a computerized family health history system. BMC Health Serv Res 2011;11:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubinstein WS, Acheson LS, O'Neill SM, et al. Clinical utility of family history for cancer screening and referral in primary care: a report from the Family Healthware Impact Trial. Genet Med 2011;13:956–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat 2011;32:532–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wells S, Kerr A, Broad J, et al. The impact of New Zealand CVD risk chart adjustments for family history and ethnicity on eligibility for treatment (PREDICT CVD-5). NZ Med J 2007;120:U2712. [PubMed] [Google Scholar]
- 52. Iavindrasana J, Lo Iacono L, Muller H, et al. The @neurIST project. Stud Health Technol Inform 2008;138:161–4 [PubMed] [Google Scholar]
- 53. Scheuner MT, De Vries H, Kim B, et al. Are electronic health records ready for genomic medicine? Genet Med 2009;11:510–17 [DOI] [PubMed] [Google Scholar]
- 54. Glasspool DW, Fox J, Coulson AS, et al. Risk assessment in genetics: a semi-quantitative approach. Stud Health Technol Inform 2001;84:459–63 [PubMed] [Google Scholar]
- 55. Emery J. Computer support for genetic advice in primary care. Br J Gen Pract 1999;49:572–5 [PMC free article] [PubMed] [Google Scholar]
- 56. Jaspers MW, Smeulers M, Vermeulen H, et al. Effects of clinical decision-support systems on practitioner performance and patient outcomes: a synthesis of high-quality systematic review findings. J Am Med Inform Assoc 2011;18:327–34 Epub 2011 Mar 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Wyk JT, van Wijk MA, Sturkenboom MC, et al. Electronic alerts versus on-demand decision support to improve dyslipidemia treatment: a cluster randomized controlled trial. Circulation 2008;117:371–8 [DOI] [PubMed] [Google Scholar]
- 58. Ullman-Cullere MH, Mathew JP. Emerging landscape of genomics in the electronic health record for personalized medicine. Hum Mutat 2011;32:512–16 [DOI] [PubMed] [Google Scholar]
- 59. Snyderman R, Yoediono Z. Prospective care: a personalized, preventative approach to medicine. Pharmacogenomics 2006;7:5–9 [DOI] [PubMed] [Google Scholar]
- 60. Samwald M, Stenzhorn H, Dumontier M, et al. Towards an interoperable information infrastructure providing decision support for genomic medicine. Stud Health Technol Inform 2011;169:165–9 [PubMed] [Google Scholar]
- 61. Hoffman MA. The genome-enabled electronic medical record. J Biomed Inform 2007;40:44–6 [DOI] [PubMed] [Google Scholar]
- 62. Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet 2011;130:33–9 [DOI] [PubMed] [Google Scholar]
- 63. Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res 2009;154:277–87 [DOI] [PubMed] [Google Scholar]
- 64. Downing GJ. Policy perspectives on the emerging pathways of personalized medicine. Dialogues Clin Neurosci 2009;11:377–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu Rev Genomics Hum Genet 2011;12:217–44 [DOI] [PubMed] [Google Scholar]
- 66. Cordero P, Ashley EA. Whole-genome sequencing in personalized therapeutics. Clin Pharmacol Ther 2012;2:51. [DOI] [PubMed] [Google Scholar]
- 67. Ali-Khan SE, Daar AS, Shuman C, et al. Whole genome scanning: resolving clinical diagnosis and management amidst complex data. Pediatr Res 2009;66:357–63 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.