Abstract
Cellulose synthase-like (CSL) genes are predicted to catalyse the biosynthesis of non-cellulosic polysaccharides such as the β-d-glycan backbone of hemicelluloses and are classified into nine subfamilies (CSLA–CSLH and CSLJ). The CSLD subfamily is conserved in all land plants, and among the nine CSL subfamilies, it shows the highest sequence similarity to the cellulose synthase genes, suggesting that it plays fundamental roles in plant development. This study presents a detailed analysis of slender leaf 1 (sle1) mutants of rice that showed rolled and narrow leaf blades and a reduction in plant height. The narrow leaf blade of sle1 was caused by reduced cell proliferation beginning at the P3 primordial stage. In addition to the size reduction of organs, sle1 mutants exhibited serious developmental defects in pollen formation, anther dehiscence, stomata formation, and cell arrangement in various tissues. Map-based cloning revealed that SLE1 encodes the OsCSLD4 protein, which was identified previously from a narrow leaf and dwarf 1 mutant. In situ hybridization experiments showed that OsCSLD4 was expressed in a patchy pattern in developing organs. Double-target in situ hybridization and quantitative RT-PCR analyses revealed that SLE1 was expressed specifically during the M-phase of the cell cycle, and suggested that the cell-cycle regulation was altered in sle1 mutants. These results suggest that the OsCSLD4 protein plays a pivotal role in the M phase to regulate cell proliferation. Further study of OsCSLD4 is expected to yield new insight into the role of hemicelluloses in plant development.
Key words: CSLD, cytokinesis, leaf blade, M-phase, rice, slender leaf 1.
Introduction
The plant cell wall plays pivotal roles such as mechanical support, forming a barrier against biotic and abiotic stresses, and control of exchanging materials and signal transduction between cells. The cell wall is composed mainly of polysaccharides, which are classified into cellulose, hemicelluloses, and pectins (Scheller and Ulvskov, 2010). Cellulose microfibrils are the core component of the cell walls, and are typically composed of approximately 36 hydrogen-bonded chains containing 500–14 000 β-1,4-linked glucose molecules (Taylor, 2008). The cellulose synthase (CesA) protein comprises 985–1088 aa, and contains a zinc finger domain at the N terminus, eight transmembrane domains, and a central cytosolic ‘catalytic’ domain known as the ‘D_D_D_QxxRW’ motif. It is considered that the CesA complex comprising 36 CesA proteins is embedded in the plasma membrane in hexameric arrays, and that the zinc finger domain at the N terminus participates in the dimerization of CesA proteins (Kurek et al., 2002; Taylor, 2008).
Richmond and Somerville (2000) reported a large family of Cellulose synthase-like (CSL) genes in Arabidopsis based on the sequence similarity to CesA genes, and classified them into six subfamilies (CSLA–E and CSLG). Subsequent studies identified three additional CSL subfamilies (CSLF, CSLH, and CSLJ) (Hazen et al., 2002; Fincher, 2009). Whilst the CSLB and CSLG subfamilies are found only in dicots, the CSLF and CSLH subfamilies are specific to grasses, and the CSLJ subfamily is found in some angiosperms. The CSLA, CSLC, and CSLD subfamilies are found in all land plants. CSL proteins contain sequence motifs that are characteristic of β-glycosyltransferases. The only difference between CSLs and CesAs is the lack of the zinc finger domains at the N terminus. Most of the CSL proteins appear to be localized in the Golgi, where hemicellulose synthesis takes place. From these characteristics, CSL genes are predicted to catalyse the biosynthesis of non-cellulosic polysaccarides such as the β-d-glycan backbone of hemicelluloses (Richmond and Somerville, 2000). This hypothesis is supported by several recent studies, which suggest that the CSLA genes encode (gluco)mannan synthases (Liepman et al., 2005; Suzuki et al., 2006), and that the CSLF and CSLH genes encode mixed-linkage glucan synthases (Burton et al., 2006; Doblin et al., 2009). Cocuron et al. (2007) suggested that the CSLC genes are involved in xyloglucan synthesis. Recently, however, it was reported that some CSLC genes of barley are targeted to the plasma membrane, suggesting that the CSLC subfamily contains more than one type of polysaccharide synthase (Dwivany et al., 2009).
The CSLD subfamily genes are commonly found in all land plants, and show the highest similarity to CesA family among CSL subfamilies at sequence levels (Richmond and Somerville, 2000). To date, six CSLD genes have been found in Arabidopsis, five in maize, and five in rice (Hunter et al., 2012), and several csld mutants were identified in these plants. In addition, homology searches have revealed that the CSLD subfamily contains three barley genes, five sorghum genes, and six purple false brome genes (Supplementary Fig. S1 at JXB online). Phylogenetic analysis revealed that CSLD genes are further classified into three clades, which correspond to three classes of their mutant phenotypes (Hunter et al., 2012). The first clade includes AtCSLD1 and AtCSLD4, whose mutants show pollen-tube defects (Bernal et al., 2008). The second clade comprises AtCSLD2, AtCSLD3, OsCSLD1, and ZmCSLD5, and their mutations cause aberrant root-hair development (Favery et al., 2001; Wang et al., 2001; Kim et al., 2007; Bernal et al., 2008; Penning et al., 2009). The third clade includes AtCSLD5, OsCSLD4, and ZmCSLD1, whose mutants show reduced plant growth (Bernal et al., 2007; Li et al., 2009; Hu et al., 2010; Wu et al., 2010; Hunter et al., 2012). Whilst expression of the first clade genes AtCSLD1 and AtCSLD4 is specific to pollens, the second clade genes AtCSLD2 and AtCSLD3 are expressed at the highest levels in root tissues except for the root tips. The third clade gene AtCSLD5 is expressed at moderate levels in many tissues including root tips and at the highest level in the shoot apex (Bernal et al., 2008). These distinct expression patterns among the three clades could correspond to different mutant phenotypes.
The reduced number of cells in atcsld5 and oscsld4 mutants and the high expression level of these genes in the meristematic tissues (Bernal et al., 2008; Li et al., 2009) have suggested a functional relation of CSLD to cell proliferation. Recently, Hunter et al. (2012) found disrupted cross-wall formation in a zmcsld1 mutant. As similar phenotypic alterations have frequently been observed in cytokinetic mutants such as knolle and korrigan in Arabidopsis (Lukowitz et al., 1996; Zuo et al., 2000), it is likely that ZmCSLD1 is associated with cytokinesis. In addition, Yin et al. (2011) revealed that transiently expressed AtCSLD5 is involved in mannan synthesis in tobacco leaves. The mannosyltransferase activity of AtCSLD5 was reduced by adding GDP-glucose together with GDP-mannose (Yin et al., 2011), suggesting that the CSLD subfamily is involved in a different kind of mannan synthesis from that catalysed by the CSLA subfamily. Although mannans have been well studied as storage components, little information has been accumulated in relation to cytokinesis.
Here, we present a detailed analysis of rice slender leaf 1 (sle1) mutants that show narrow leaf blades and other developmental abnormalities, the phenotypes being attributable to reduced cell proliferation activity. Map-based cloning revealed that SLE1 encodes OsCSLD4. This gene is the same as the previously reported NARROW LEAF AND DWARF 1 (ND1) gene (Li et al., 2009). The nd1 mutant showed narrow leaves and reduced plant height. Li et al. (2009) revealed the localization of the OsCSLD4 protein in the Golgi. In the present study, we found novel mutant phenotypes that were not described by Li et al. (2009), and revealed that OsCSLD4/SLE1 is specifically expressed during the M phase of the cell cycle. These results suggest that the product of OsCSLD4 plays a pivotal role in the M phase to regulate cell proliferation and plant development.
Materials and methods
Plant materials
Two allelic single-gene recessive mutants of rice (Oryza sativa L.) showing distinct rolled and narrow leaf blades and reduced plant height were identified from an M2 population of japonica variety Taichung 65 mutagenized with N-methyl-N-nitrosourea. We designated these mutants slender leaf1-1 (sle1-1) and sle1-2, respectively. For observations at the early vegetative stage, mutants and wild-type seeds were sown on soil and grown at 28 °C under continuous light conditions. Otherwise, plants were grown in pots or in paddy fields under natural conditions.
Epidermal cell observations
The third-leaf blades of sle1 and wild-type plants were fixed with formaldehyde:glacial acetic acid:50% ethanol (2:1:17) for 24h at 4 °C. They were then dehydrated in a graded ethanol series. Dehydrated samples were incubated at 96 °C in chloral hydrate dissolved in 100% ethanol until they cleared and were then observed under a light microscope.
Paraffin sectioning and histological analysis
Plant samples of sle1 and wild-type were fixed with formaldehyde:glacial acetic acid:50% ethanol (2:1:17)) for 24h at 4 °C for histological analysis, or fixed with 4% (w/v) paraformaldehyde and 1% Triton X in 0.1M sodium phosphate buffer for 48h at 4 °C for in situ hybridization. They were then dehydrated in a graded ethanol series, substituted with 1-butanol, and embedded in Paraplast Plus. The samples were sectioned at 8 μm thick using a rotary microtome. For histological analysis, the sections were stained with haematoxylin or calcofluor and observed under a light or fluorescence microscope.
Flow cytometric analysis
Mature leaves of wild-type, sle1-1, and sle1-2 plants were cut into small pieces in 430 μl of extraction buffer (CyStain® UV Precise P), filtered through a 20 μm mesh, and mixed with 1.6ml of nuclear staining buffer (Partec). The samples were analysed with a Partec Ploidy Analyzer (Partec). For quantification, the mean result of three independent measurements was determined.
In situ hybridization
Paraffin sections were prepared as described above. Digoxigenin-labelled antisense and sense RNA probes were prepared from full-length cDNA of histone H4 and a 752bp fragment of OsCSLD4/SLE1, which was amplified by PCR with forward primer (5′-CAGGGCCTACTCAAGGTGAT-3′) and reverse primer (5′-GCACAGACGTCACACACACA-3′) using genomic DNA as a template. Biotin-labelled antisense and sense RNA probes were prepared from full-length cDNA of CDKB2;1. As the sense probes did not give specific signals, only antisense probe data are presented. For single-target in situ hybridization, digoxigenin-labelled probes were used. In situ hybridization and immunological detection with alkaline phosphatase were performed according to the methods of Kouchi and Hata (1993). The measurement of histone H4 signal areas in leaf primordia was performed by image analysis with ImageJ (http://rsbweb.nih.gov/ij/). For double-target in situ hybridization, a digoxigenin-labelled probe and biotin-labelled probe were used. Hybridization of probes, post-hybridization washes, and blocking procedures were performed according to the methods of Kouchi and Hata (1993). A TSATM Biotin System (PerkinElmer) together with streptavidin–fluorescein (PerkinElmer) was used for the detection of biotin-labelled probes and an HNPP Fluorescent Detection Set (Roche) was used for the detection of digoxigenin-labelled probes following the manufacturer’s protocol. The slides were washed in sterile distilled water and mounted with Prolong Gold Antifade Reagent with DAPI (Invitrogen), and observed with a BZ-8000 fluorescence microscope (Keyence Co.) using GFP band pass, Texas Red, and DAPI band pass filters for fluorescein, HNP/TR, and DAPI signals, respectively.
Map-based cloning
For mapping of the SLE1 gene, sle1-1 and sle1-2 plants were crossed with indica variety Kasalath, and F2 plants showing the sle1 phenotype were used for mapping. Using cleaved-amplified polymorphic sequence and sequence-tagged site markers, the SLE1 locus was mapped roughly on the long arm of chromosome 12. The SLE1 locus was further limited within the region ranged from 78.9 to 91.9 cM. As OsCSLD4 (Os12g36890) located in this region was reported as a causal gene for the nd1 mutant of rice (Li et al., 2009), we compared the genomic sequence of the gene between sle1 mutants and wild-type plants.
Quantitative RT-PCR analysis
Total RNA was extracted from whole seedlings using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. After RNase-free DNase (Takara Bio) treatment, 500ng of total RNA was reverse transcribed with a High Capacity RNA-to-cDNATM Master Mix (Applied Biosystems) according to the manufacturer’s protocol. Quantitative RT-PCR analyses of histone H4, CDKB2;1, and UBQ5 were performed using Fast SYBR® Green Master Mix (Applied Biosystems) and quantitative RT-PCR analyses of SLE1 and eEF-1a were performed using TaqMan® Fast Universal PCR Master Mix (Applied Biosystems). Primer sequences used for quantitative RT-PCR were as follows; 5′-CTGATTCCTTCCTTCCTTCCTT-3′ and 5′-CAAATCTTCAGTTCAGATTCATCG-3′ for histone H4, 5′-TGATGTTCACCACCAAGGAA-3′ and 5′-TCATTACCATCA CAAACAAGTGAA-3′ for CDKB2;1, 5′-ACCACTTCGACCGC CACTACT-3′ and 5′-ACGCCTAAGCCTGCTGGTT-3′ for UBQ5, 5′-GAAGCTCACATGAAGGAGTGC-3′ and 5′-CAAACAATCG CTTAATCACACC-3′ for OsCSLD4/SLE1, and 5′-CGCTCTTCTT GCTTTCACTCTTG-3′ and 5′-TAGGATGAGACTTCCTTCACG ATTTC-3′ for eEF-1a. The probe sequences used for quantitative RT-PCR were as follows; 5′-GGCGTGTTGTCAAATTCAGTT-3′ for SLE1 and 5′-CAACAAGATGGATGCCAC-3′ for eEF-1a. The SEL1 probe was labelled with FAM and TAMRA, and the eEF-1a probe was labelled with FAM and NFQ-MGB. Quantitative RT-PCR analysis was performed using the StepOnePlusTM Real-Time PCR System (Applied Biosystems).
Results
We identified two alleles of SLE1: sle1-1 and sle1-2. As several phenotypes of SLE1 have already been described by Li et al. (2009), we focused on a detailed observation of the developmental process in the mutant. We also found an abnormality in the reproductive phase that was not described by Li et al. (2009).
Phenotypes of sle1 mutants in the vegetative phase
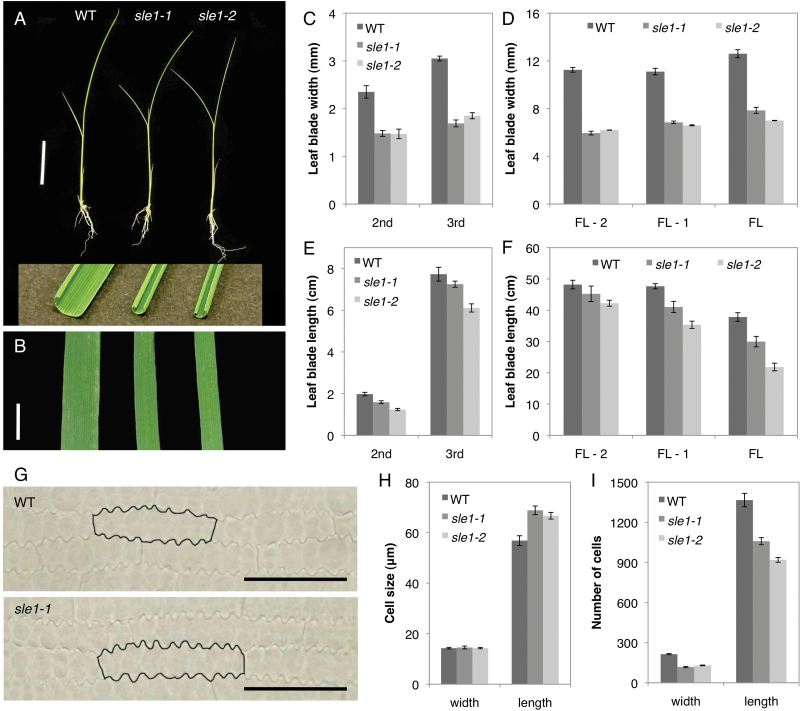
Both sle1-1 and sle1-2 showed rolled and narrow leaf blades (Fig. 1A, B). The width of the leaf blade was reduced to about 60% of that of the wild type, and this trend was observed in all leaves (Fig. 1C, D). The length of the leaf blade was also reduced, although it was less severe than the reduction in width (Fig. 1E, F). To compare the epidermal cells in the leaf blades, we cleared the third leaves of wild-type and sle1 mutant plants with chloral hydrate (Fig. 1G). On the abaxial surface, no significant difference was observed in the width of the intercostal epidermal cells, but the length of the intercostal epidermal cells in the sle1 mutants was 20% greater than in the wild type (Fig. 1H). A similar trend was also observed in the cork cells of costal cells in the third-leaf blades (Supplementary Fig. S2 at JXB online). In the sle1 mutant, the total number of cells along the leaf width was reduced by approximately 40%, and along the leaf length, it was reduced by approximately 30% compared with the wild type (Fig. 1I). Thus, the short and narrow leaves of sle1 mutants were caused by a defect in cell proliferation. Concomitant with the leaf size, sle1 mutants also showed a dwarf phenotype at maturity due to a reduction in the lengths of the two uppermost internodes (Supplementary Fig. S3 at JXB online). Abnormalities were also observed in the belowground plant parts. The roots of the sle1 mutants were more slender and shorter compared with those of the wild type, and the number of crown roots was decreased in the mutants (Supplementary Fig. S4 at JXB online). The sle1 mutants also exhibited poor development of lateral roots and root hairs (Supplementary Fig. S4).
Fig. 1.
Vegetative phenotypes of wild-type, sle1-1, and sle1-2 plants. (A) Fourteen-d-old seedlings of wild-type, sle1-1, and sle1-2 plants. Sections of the third-leaf blades are shown in the inset. (B) Mechanically unrolled third-leaf blades of wild-type, sle1-1, and sle1-2 plants (left to right). Bars, 5cm (A), 3mm (B). (C–F) Comparison of width (C, D) and length (E, F) of leaf blades among wild-type, sle1-1, and sle1-2 plants. Results for the second and third leaves are shown, and for the flag leaf (FL) and leaves one and two lower than the flag leaf (FL-1 and FL-2, respectively). Results are shown as means ±standard error (SE) (n=10). (G) The abaxial surface of the third-leaf blade of wild-type and sle1-1 plants. The representative cells are outlined in black. Bars, 50 μm. (H, I) Comparison of epidermal cell size (H) and number of cells (I) in the third-leaf blade among wild-type, sle1-1, and sle1-2 plants. To estimate the number of cells comprising the third-leaf blade, the width and length of the leaf blade were divided by those of epidermal cells. Results are shown as means ±SE (n=5). (This figure is available in colour at JXB online.)
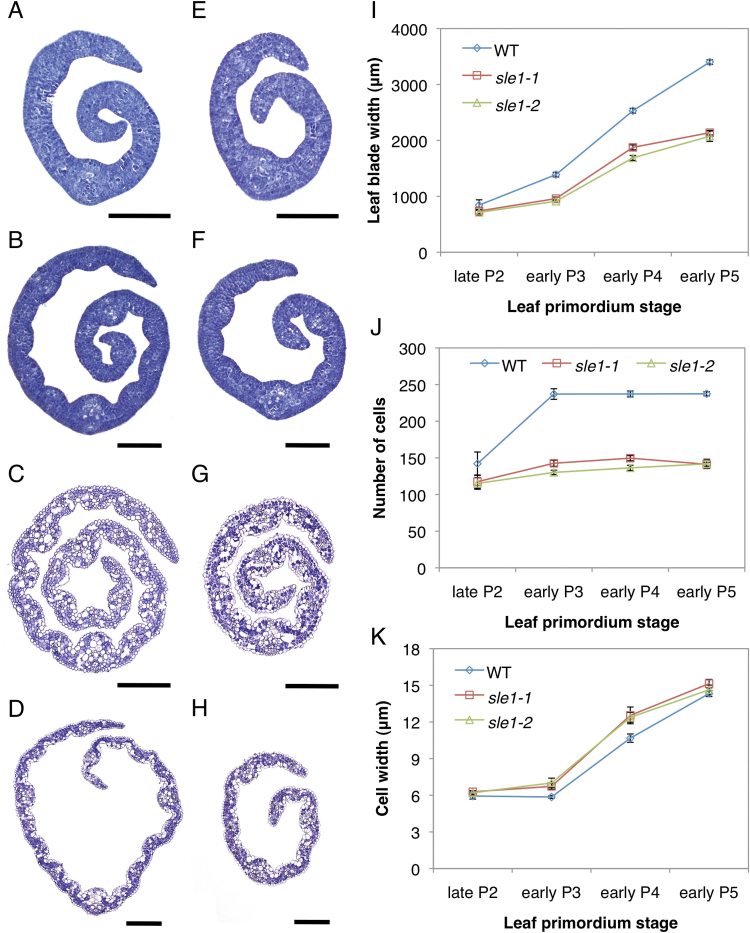
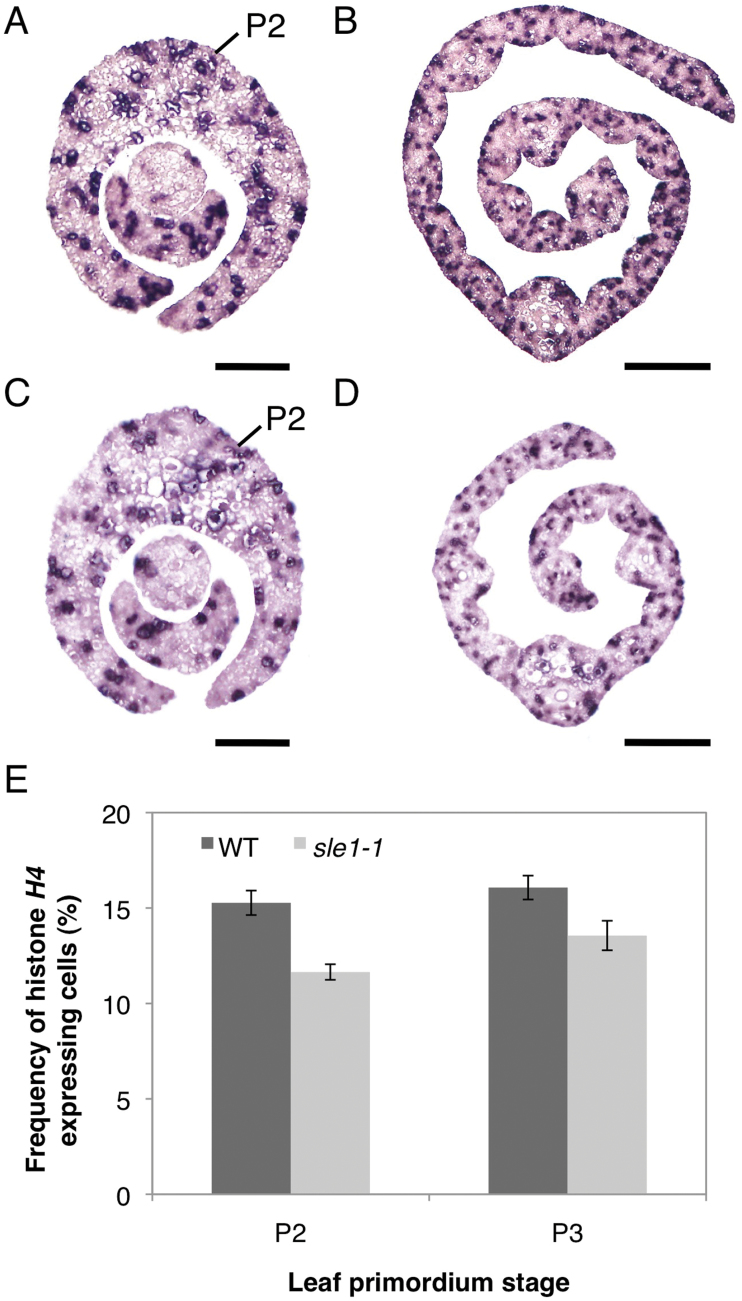
We investigated the early leaf development of sle1 plants to determine when leaf reduction begins in the mutant. Although the width of the late P2 leaf primordium in sle1 plants was comparable to that of the wild type (Fig. 2A, E, I), sle1 plants already exhibited narrower leaf blades in the early P3–P5 leaf primordia (Fig. 2B–D, F–I). To determine the cause of leaf narrowness in sle1 plants, we estimated the number of epidermal cells in the width direction of the third-leaf blade. Although the number of cells did not largely differ at the late P2 stage, sle1 plants showed significantly fewer cells from the early P3 stage (Fig. 2J). On the other hand, epidermal cell width did not largely differ between sle1 and wild-type plants (Fig. 2K). These results indicated that the narrow leaves of sle1 mutants are caused mainly by impaired cell proliferation. To confirm the impaired cell proliferation in sle1 mutants, we used in situ hybridization to examine the expression of histone H4, which is expressed specifically in S-phase cells (Fig. 3A–D). For quantitative evaluation, the relative number of cells expressing histone H4 was calculated using ImageJ software. The frequency of cells expressing histone H4 was reduced in both P2 and P3 leaf primordia in sle1-1 plants (Fig. 3E) relative to the wild type, indicating that the narrow leaf blade of sle1 plants is caused by impaired cell proliferation at an early stage.
Fig. 2.
Development of the third-leaf blade in wild-type, sle1-1, and sle1-2 plants. (A, E) Cross-sections of late P2 leaf blades in wild-type (A) and sle1-1 (E) plants. (B, F) Cross-sections of early P3 leaf blades in wild-type (B) and sle1-1 (F) plants. (C, G) Cross-sections of early P4 leaf blades in wild-type (C) and sle1-1 (G) plants. (D, H) Cross-sections of early P5 leaf blades in wild-type (D) and sle1-1 (H) plants. Bars, 100 μm (A, B, E, F); 200 μm (C, D, G, H). (I) Increase in the third-leaf blade width at the early stages in wild-type, sle1-1 and sle1-2 plants. (J) Number of epidermal cells in the width direction at the early stages in the third-leaf blade. (K) Epidermal cell width at the early stages in the third-leaf blade. Leaf blade width was calculated from the cross-sections of leaf primordium at each stage using ImageJ software. The widths of epidermal cells in late P2 and early P3 leaf primordium were calculated from the longitudinal sections of leaf blade, and those in early P4 and P5 leaf primordium were calculated from the cleared leaf blade (see Materials and methods) using ImageJ software. To estimate the number of cells, the leaf blade width was divided by the mean epidermal cell width. Results are shown as means ±SE (n=3). (This figure is available in colour at JXB online.)
Fig. 3.
Expression patterns of histone H4 in P2 and P3 leaf primordia in wild-type and sle1-1 plants. Ten-d-old seedlings were used. (A, C) Cross-sections of P2 leaf primordia in wild-type (A) and sle1-1 (C) plants. (B, D) Cross-sections of a P3 leaf blade in wild-type (B) and sle1-1 (D) plants. Bars, 50 μm (A, C); 100 μm (B, D). (E) Frequency of histone H4-expressing cells in P2 and P3 leaf primordia. Results are shown as means ±SE (n=6). (This figure is available in colour at JXB online.)
Phenotypes of sle1 mutants in the reproductive phase
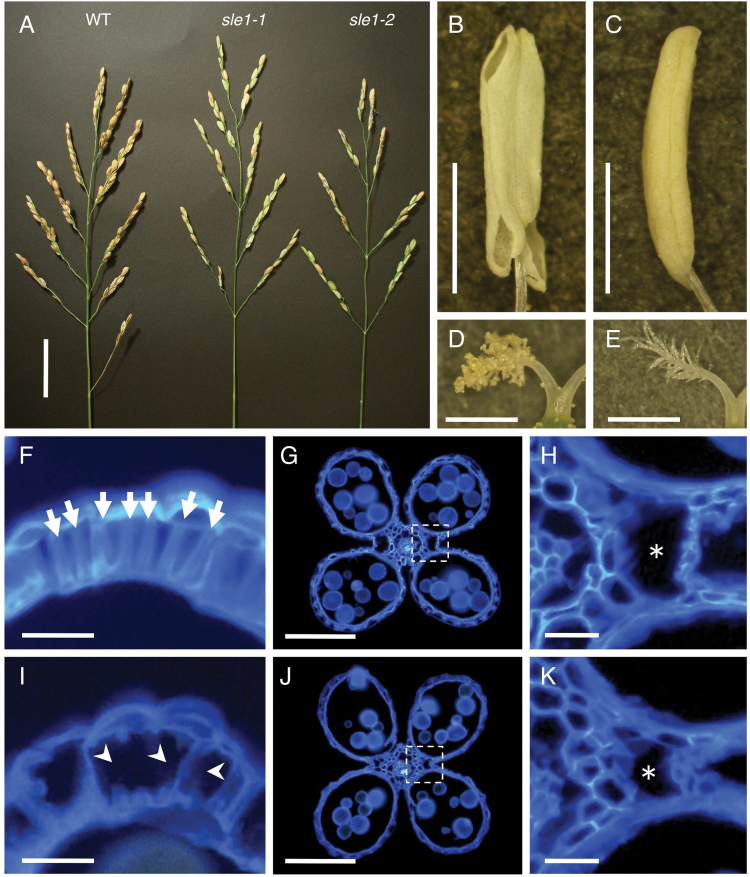
The sle1 mutants showed distinct abnormalities at the reproductive stage, which have not been described in detail previously. The panicles of sle1 mutants were shorter than those of the wild type (Fig. 4A, Table 1). The number of spikelets per panicle was significantly reduced due primarily to a decreased number of primary branches (Table 1). The slender leaf phenotype was also reflected in the spikelets; spikelet width in sle1 plants was significantly reduced, while the spikelet length was comparable to that of wild type (Table 1). Presumably, these reductions in both the number and width of spikelets can be attributed to the defect in cell proliferation. One of the most striking features of the sle1 panicles was low seed fertility (approximately 30% in sle1 plants compared with >90% in the wild type). Therefore, many of the spikelets remained green when the seeds matured (Fig. 4A). To discover the reason for this low fertility, we first observed the ovules of sle1 mutants, but no significant phenotypic alteration was detected. Furthermore, sle1 plants could set seeds when crossed with wild-type plants, eliminating the pistils as the cause of low fertility. We then examined the dehiscence of the anthers. In wild-type plants, dehiscence occurred at flower opening and the stigmas were covered with many pollen grains (Fig. 4B, D), whereas in sle1 mutants, few anthers dehisced and few pollen grains was observed on the stigmas (Fig. 4C, E). Therefore, poor dehiscence of anthers appeared to be the major cause of low seed fertility in the sle1 mutants. The driving force for anther dehiscence normally arises from fibrous structures in the endothecium cells, along with the development of cavities and the rupture of septa that are indispensable for dehiscence (Matsui et al., 1999). In the sle1 mutants, development of both the fibrous structures in the endothecium cells and the necessary cavities for dehiscence was incomplete (Fig. 4F–K). Thus, the SLE1 gene is also involved in the development of these structures.
Fig. 4.
Reproductive phenotypes of wild-type, sle1-1, and sle1-2 plants. (A) Panicles of wild-type, sle1-1, and sle1-2 plants. (B–E) Anthers and stigmas of wild-type (B, D) and sle1-1 (C, E) plants at flower opening. The anthers were well dehisced in wild-type plants (B), but anther dehiscience was hardly observed in sle1 plants (C). Numerous pollen grains are attached to the stigmas in wild-type plants (D), with only a few attached on the sle1 stigma (E). (F–K) Cross-sections of anthers in wild-type (F, G) and sle1-1 (I, J) plants stained with calcofluor. (F) and (I) are sections of the upper part of the anther. In (F), the arrows indicate normal fibrous structures formed in endothecium cells. In (I), the fibrous structures in the endothecium cells were partly lost (arrowheads) in sle1-1 plants. (G) and (J) are sections of the middle part of the anther. (H) and (K) show close-up views of the boxed regions in (G) and (J), respectively. Asterisks indicate cavities for dehiscence. Bars, 5cm (A); 1mm (B–E); 10 μm (F, H, I, K); 100 μm (G, J).
Table 1.
Summary of panicle traits in wild-type and sle1 plants. Values indicate the mean of ten samples ±SE. Asterisks indicate results that were significantly different from the wild type (WT) at P <0.05 (*) and P <0.01 (**) (t-test).
| Panicle length (cm) | No. of primary branches per panicle | No. of spikelets per panicle | Spikelet length (mm) | Spikelet width (mm) | |
|---|---|---|---|---|---|
| WT | 21.9±0.7 | 10.8±0.4 | 114.3±8.8 | 7.6±0.1 | 3.7±0.05 |
| sle1-1 | 18.9±0.5** | 7.0±0.6** | 57.5±4.5** | 7.7±0.2 | 2.9±0.04** |
| sle1-2 | 15.1±0.6** | 6.4±0.4** | 40.0±3.2** | 8.0±0.1* | 3.1±0.03** |
The sle1 mutant is defective in cell division and cell size/shape regulation
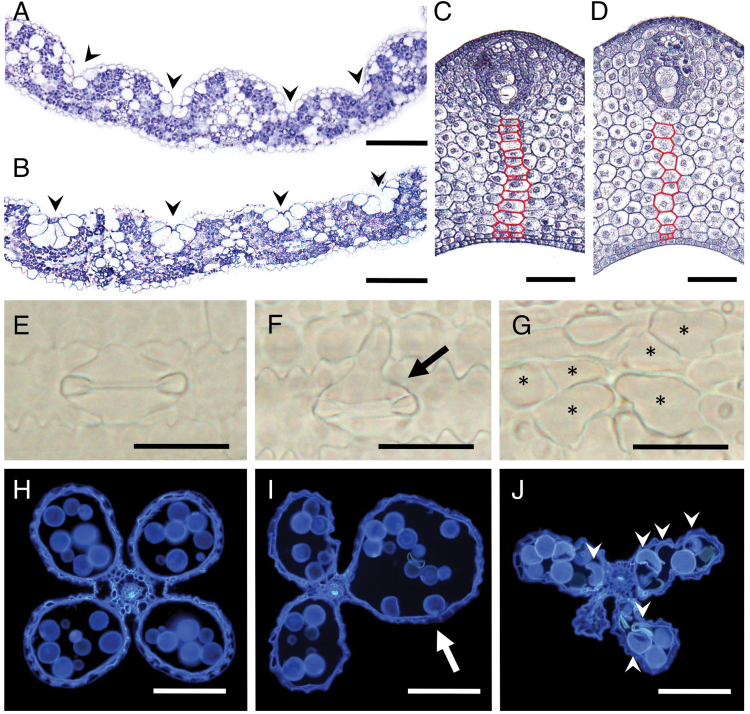
The sle1 mutants showed pleiotropic abnormalities in pollen formation, anther dehiscence, stomata formation, and cell arrangement in various tissues, as well as altered organ size. In the leaf blades, bulliform cells and bundle-sheath cells were enlarged, but the vascular bundles were smaller than those of the wild type (Fig. 5A, B). In addition, the number of vascular bundles in the leaf blades was significantly reduced in the sle1 mutants, whilst the distance between vascular bundles was not affected (Supplementary Fig. S5 at JXB online). In the leaf sheath of the sle1 mutant, the cells were apparently enlarged, together with a reduction in cell number (Fig. 5C, D). The stomata were sometimes ill formed (Fig. 5E, F), and occasionally enlarged cells of unknown identity were observed in the stomatal lineage (Fig. 5G). As these cells were always positioned where the stomata should have been formed, these cells appeared to be abnormal stomata cells, suggesting that improper cell divisions may have taken place in the course of stomata development. Occasionally, anthers showed abnormal development, and many of the pollen grains in such anthers were also misshapen in the sle1 mutants (Fig. 5H–J). Thus, the SLE1 mutation affected not only the cell number and size but also the cell shape and tissue differentiation.
Fig. 5.
Abnormal cell division and wide variation in cell size/shape in sle1 tissues. (A, B) Cross-sections of the third-leaf blades in wild-type (A) and sle1-1 (B) plants. Arrowheads indicate the positions of bulliform cells. (C, D) Cross-sections of the leaf sheath of P4 leaf primordia in wild-type (C) and sle1-2 (D) plants. Along the representative cell file (indicated in red) extending from the vascular bundle to the adaxial epidermis, there were 16 cells in the wild-type plant, but only ten cells in the sle1-2 plant. (E–G) Stomata in wild-type (E) and sle1-2 (F) plants. The arrow in (F) indicates an abnormal stomata. The asterisks in (G) indicate enlarged cells at the position where stomata should be formed. (H–J) Cross-sections of anthers in wild-type (H), sle1-1 (I), and sle1-2 (J) plants. The arrow in (I) indicates an abnormal pollen sac and the arrow heads in (J) indicate defective pollen grains. Bars, 100 μm (A, B, H–J); 50 μm (C, D); 25 μm (E–G).
The SLE1 gene encodes OsCSLD4
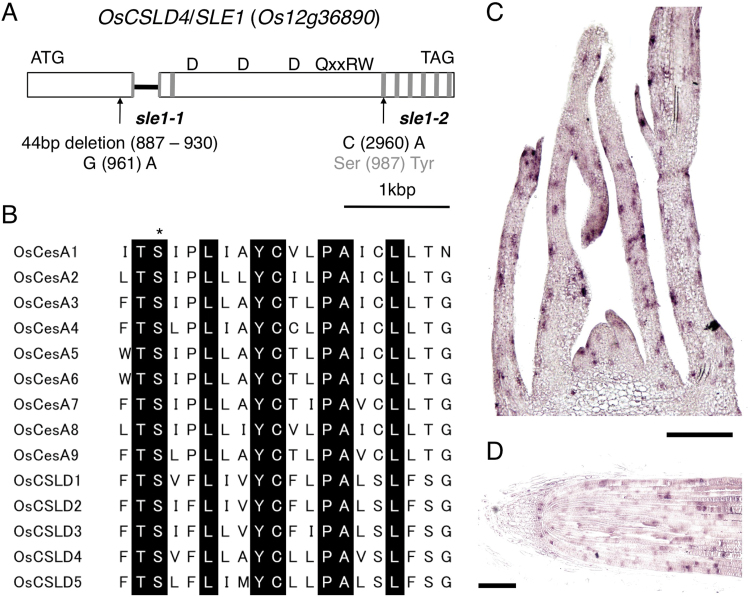
Map-based cloning using an F2 population from sle1-1, sle1-2, and ssp. indica variety Kasalath revealed that SLE1 encodes OsCSLD4, which is identical to the previously reported ND1 protein (Li et al., 2009) (Fig. 6A). The OsCSLD4 protein contains a ‘D_D_D_QxxRW’ motif with two and six transmembrane domains at the N- and C-terminal regions, respectively. In sle1-1, a 44bp fragment ranging from nt 887 to 930 was deleted, which caused a frameshift with concomitant loss of function of all of the transmembrane domains and central motif. In addition to the above deletion, a single-nucleotide substitution from G to A at nt 961 occurred in sle1-1 plants. In sle1-2 plants, a nucleotide was substituted from C to A at nt 2960, which caused an amino acid substitution from Ser to Tyr in the third transmembrane domain from the N terminus. This Ser is conserved throughout CesA and all CSLD proteins in rice (Fig. 6B). Therefore, this amino acid substitution conceivably could impair the function of this transmembrane domain and affect the function of the OsCSLD4 protein.
Fig. 6.
Structure of the OsCSLD4/SLE1 gene, alignment of the third transmembrane domain from the N terminus of CesA and CSLD genes in rice, and expression pattern of OsCSLD4. (A) Structure of the OsCSLD4 gene (Os12g36890). Open boxes represent exons, and the filled bar represents the intron. Transmembrane domains are represented by grey boxes (the first transmembrane domain from the N terminus is split by the intron). The central domain is represented by ‘D_D_D_QxxRW’. Arrows indicate the positions of the sle1-1 and sle1-2 mutations, respectively. (B) Alignment of amino acid sequences in the third transmenbrane domains in rice CesA and CSLD proteins. Amino acids conserved in all genes are represented by black boxes. The mutated residue in the sle1-2 mutant is indicated by an asterisk. (C, D) In situ hybridization of OsCSLD4 in longitudinal sections of 10-d-old shoot apex (C) and root tip (D). Bars, 100 μm. (This figure is available in colour at JXB online.)
OsCSLD4/SLE1 is expressed during the M phase of the cell cycle
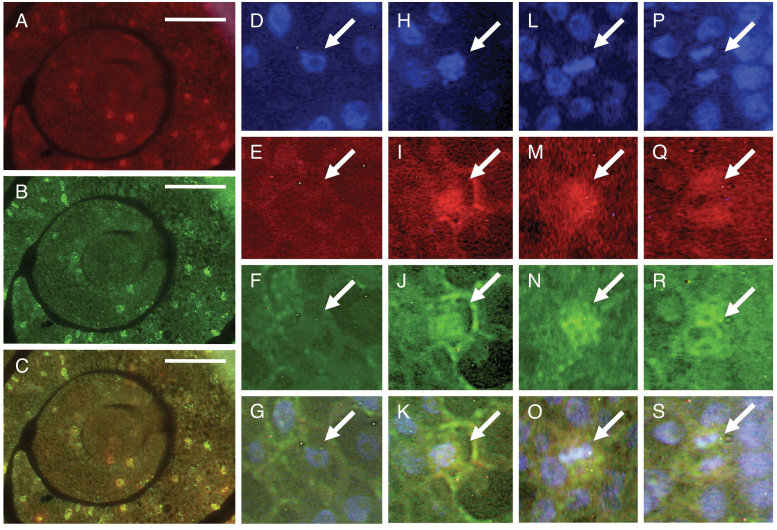
Using RT-PCR and a promoter–β-glucuronidase construct, Li et al. (2009) showed that OsCSLD4/SLE1 was expressed primarily in rapidly growing tissues. To determine cell types in which OsCSLD4 is expressed, we examined the detailed expression pattern of OsCSLD4 by in situ hybridization. No signals were detected when we used a sense probe of OsCSLD4, so only results obtained using an antisense probe are described here. The OsCSLD4 transcripts were detected in actively developing tissues, such as the shoot apex and root tip (Fig. 6C, D), in agreement with Li et al. (2009). However, the expression in any given tissue was not uniform but patchy; i.e. within a tissue, the OsCSLD4-expressing cells were dispersed (Fig. 6C, D). This patchy expression pattern suggests that OsCSLD4 expression is associated with the cell cycle, similar to the S-phase-specific histone H4 or the (G2-)M-phase-specific CDKB2;1 (Umeda et al., 1999). Therefore, we performed a double-target in situ hybridization of OsCSLD4 counterstained with histone H4 or CDKB2;1. Cells expressing OsCSLD4 did not coincide with those expressing histone H4 (Supplementary Fig. S6 at JXB online), but coincided with those expressing CDKB2;1 (Fig. 7A–C), indicating that OsCSLD4 is expressed in M-phase cells. The signals of OsCSLD4 could be observed in prophase, metaphase, and anaphase cells (Fig. 7D–S), suggesting that OsCSLD4 was expressed throughout the M phase.
Fig. 7.
Double-target in situ hybridization of OsCSLD4 and CDKB2;1 in the shoot apex of wild-type plants. Ten-d-old seedlings were used. (A–C) Cross-sections of the shoot apex. Bars, 50 μm. (D–S) Results are shown for interphase (D–G), prophase (H–K), metaphase (L–O), and anaphase (P–S) cells, respectively. Arrows indicate the representative cells. Expression patterns of OsCSLD4 (A, E, I, M, Q) and CDKB2;1 (B, F, J, N, R) are shown, together with DAPI staining (D, H, L, P). Merged views of (A and B), (D–F), (H–J), (L–N), and (P–R) are shown in (C), (G), (K), (O), and (S) respectively.
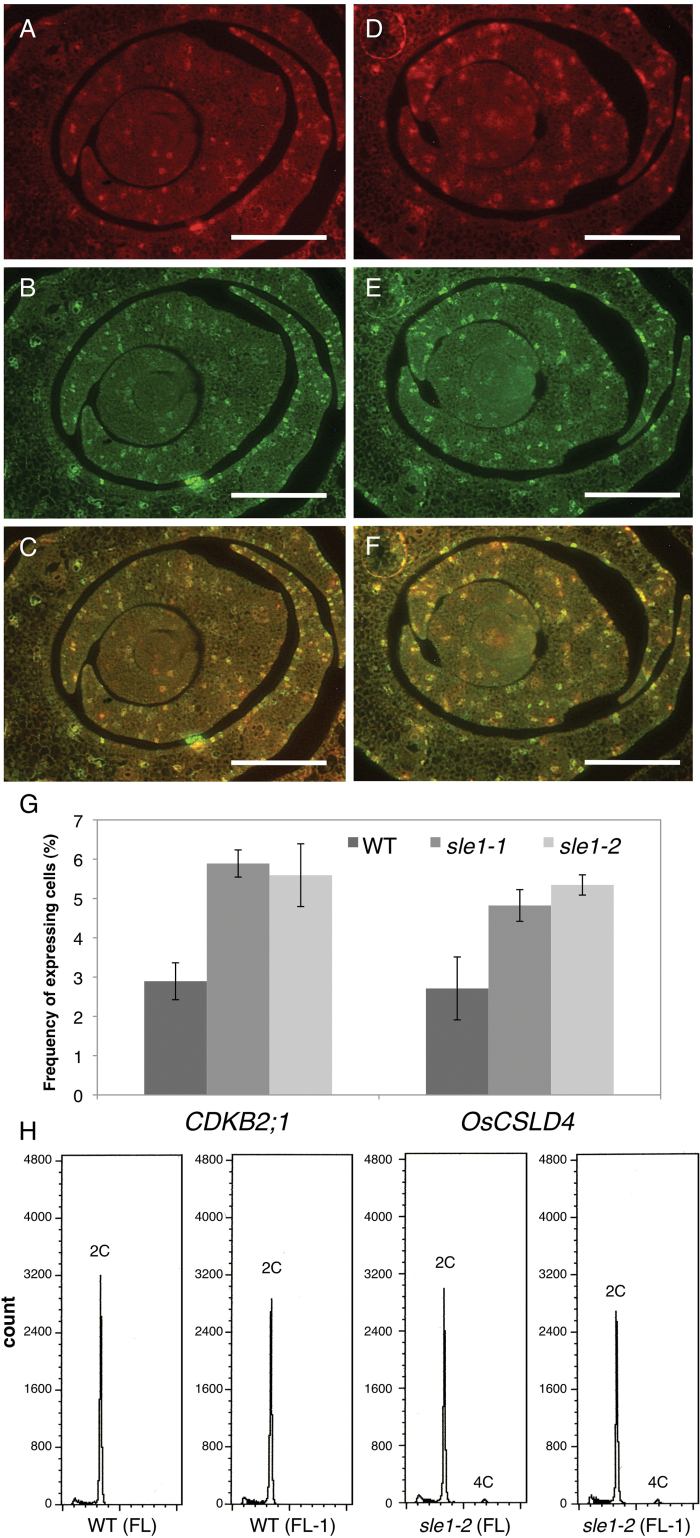
Next, we re-evaluated the mutant phenotypes in light of the observed specific expression of OsCSLD4 in M-phase cells. In Fig. 3, we showed decreased expression of histone H4 in sle1 mutants, suggesting a reduced frequency of S-phase cells. Therefore, we examined the expression of CDKB2;1 and OsCSLD4 in the sle1 mutants. OsCSLD4 signals were detected in sle1 cells expressing CDKB2;1, indicating that the M-phase specificity of OsCSLD4 expression was not altered in the sle1 mutants (Fig. 8A–F). However, the number of signals of both OsCSLD4 and CDKB2;1 increased in the sle1 mutants (Fig. 8G). To quantify gene expression, we performed quantitative RT-PCR analyses of histone H4, CDKB2;1, and OsCSLD4 in wild-type and sle1 plants (Supplementary Fig. S7 in JXB online). The expression levels of histone H4 were reduced in sle1 plants, in agreement with Fig. 3. However, the expression of CDKB2;1 and OsCSLD4 was significantly enhanced in the sle1 mutants, in agreement with Fig. 8A–G. These results indicated an altered cell-cycle regulation in sle1 mutants, which could be responsible for the reduced cell proliferation and enlarged cell size in these plants.
Fig. 8.
Expression of OsCLSD4/SLE1 and CDKB2;1, and measurement of DNA content in sle1 plants. (A–F) Double-target in situ hybridization of OsCLSD4/SLE1 counterstained with CDKB2;1 in cross-sections of wild-type and sle1-2 shoot apices. Ten-d-old seedlings were used. (A–C) Wild-type plants. (D–F) sle1-2 plants. (A, D) Expression pattern of OsCSLD4. (B, E) Expression pattern of CDKB2;1. (C, F) Merged views of (A and B) and (D and E), respectively. Bars, 100 μm. (G) Frequency of CDKB2;1- and OsCSLD4-expressing cells in P2 leaf primordia. Results are shown as means ±SE (n=3). (H) Flow cytometric measurement of DNA content in mature leaf-blade cells of wild-type and sle1-2 plants. FL and FL-1 indicate the flag leaf and one leaf lower than the flag leaf, respectively.
The above results suggest the possibility that cytokinesis is affected in the sle1 mutant, and thus that sle1 mutant has defects in cell division. Therefore, we measured the DNA content in mature leaf blade cells by flow cytometry. All the wild-type leaf-blade cells had 2C-DNA (diploid). In contrast, although most of sle1 cells had 2C-DNA, a small number of sle1 cells showed 4C-DNA (tetraploid; Fig. 8H). These tetraploid cells suggested that incomplete cytokinesis may take place in sle1 mutants.
Therefore, we concluded that OsCSLD4/SLE1 plays a pivotal role in cell proliferation and plant development via regulation of M-phase progression.
Discussion
We identified the causal gene of the sle1 mutation, which causes pleiotropic abnormalities such as narrow leaves, as OsCSLD4, which was reported previously by Li et al. (2009). The OsCSLD4 gene was expressed specifically during the M phase of the cell cycle (Fig. 7), and the sle1 mutation in OsCSLD4 apparently caused an alteration in cell-cycle regulation: the frequency of S-phase cells was reduced in sle1 mutants in agreement with the impaired cell proliferation, whereas the frequency of M-phase cells was increased significantly (Fig. 3, Fig. 8A–G, Supplementary Fig. S7). Therefore, these results implied that progression of the M phase is disturbed by the loss of SLE1 function, and the prolonged M phase resulted in a prolonged cell cycle, which would be responsible for the impaired cell proliferation and relatively short S phase in sle1 mutants. In addition, the sle1 mutation affected not only the cell number and size but also the cell shape, and tissue differentiation (Fig. 5). Thus, OsCSLD4 plays a pivotal role in plant development.
Several studies have suggested a conserved function of CSLD genes in plants, and reduced cell proliferation has been reported in atcsld5/oscsld4/zmcsld1 mutants (Bernal et al., 2007; Li et al., 2009; Hu et al., 2010; Wu et al., 2010; Hunter et al., 2012). In this study, we suggest that the reduced cell proliferation might be attributable to the prolonged M phase in sle1 mutants. Similar results were obtained in a maize zmcsld1 mutant in which cells with 4C nuclei were detected with high frequency (Hunter et al., 2012). In a preliminary examination of the expression pattern of AtCSLD5, an Arabidopsis orthologue of OsCSLD4, we saw by in situ hybridization that AtCSLD5 was expressed in developing organs in a similar patchy pattern as OsCSLD4 (Supplementary Fig. S8). Menges et al. (2003) performed microarray analysis using synchronized cell suspensions of Arabidopsis and suggested that AtCSLD5 expression occurred at M phase. Therefore, it is considered that the molecular function of the CSLD genes is conserved among rice, maize, and Arabidopsis.
Malformations of cells were typically observed in the stomata formation in sle1 plants (Fig. 5E–G). Stomata are normally formed by three highly coordinated cell divisions after the establishment of stomatal cell files in immature leaves (Kamiya et al., 2003). Malformations of stomata have also been observed in cytokinetic mutants (Söllner et al., 2002). As the cell-cycle regulation is impaired in sle1 mutants, it is plausible that the abnormal stomata in sle1 leaves resulted from improper timing of cell division in the course of stomata development. We also observed abnormally enlarged and malformed cells in the leaves and anthers of sle1 (Fig. 5A–D, H–J). A zmcsld1 mutant was shown to have defective cross-wall formation (Hunter et al., 2012). Such phenotypic alterations have been observed frequently in cytokinetic mutants of Arabidopsis, such as knolle, korrigan, and hinkel (Lukowitz et al., 1996; Zuo et al., 2000; Strompen et al., 2002). In these mutants, impairment of cytokinesis was often caused by a failure of cell-plate formation. Thus, we speculate that failure of cell-plate formation may have caused the abnormal cells that we observed in the sle1 mutants. This proposition is supported by the presence of 4C cells in the sle1 leaf blade (Fig. 8H). Although the frequency of cells containing 4C-DNA was low, as endoduplication does not occur in normal rice leaf blade, it is probable that in sle1 leaf blade, impaired cell-plate formation eventually results in the failure of cytokinesis and formation of a 4C nucleus by nuclear fusion (Klindworth and Williams, 2001).
During cytokinesis, Golgi-derived vesicles containing cell-wall materials are transported to the equatorial zone, where they fuse rapidly from the centre of the equatorial zone to form the cell plate (Samuels et al., 1995). The CSLD subfamily is considered to function in the synthesis of polysaccharides, particularly mannan (Yin et al., 2011), and the OsCSLD4/AtCSLD5 proteins are localized in the Golgi apparatus (Bernal et al., 2007; Li et al., 2009). These observations, along with our results that OsCSLD4 is expressed specifically in M-phase cells, suggest the possibility that OsCSLD4 is involved in cell-plate formation.
In addition to a reduction in leaf size and plant height, we also observed poor dehiscence of anthers in sle1 plants (Fig. 4B–E), mainly due to the poor development of fibrous structures in the endothecium cells and underdeveloped cavities for dehiscence (Fig. 4F–K). Aberrant anthers were observed occasionally in the sle1 mutants, and many of the pollen grains in these anthers were defective (Fig. 5H–J). Similar phenotypic alterations in anthers and pollen grains are caused by environmental stresses such as drought stress (Ji et al., 2011). Rice is the most drought-sensitive plant among the major cereal crops, and reproductive development is especially susceptible to drought stress. Because the sle1 mutants exhibited a distinct rolled-leaf phenotype, which is a common dehydration-avoidance mechanism in rice, the sle1 mutants may also have experienced drought stress to some extent, presumably because of the impaired development of roots (Supplementary Fig. S4). Another possible mechanism for the defective anthers and pollen grains in sle1 plants is an abnormality in meiosis. Zhou et al. (2011) described a pollen semi-sterility1 (pss1) mutant, which displayed defective anther dehiscence and reduced pollen viability due to defective male meiosis. The phenotypic similarity between sle1 and pss1 also suggests the possibility of an impairment in meiosis in the sle1 mutant. From these phenotypic alterations, sle1 is clearly involved not only in cell proliferation but also in proper plant development.
The reduction in leaf-blade width was attributable to the reduction in cell number in the sle1 mutants (Fig. 1I). The narrow leaf blade phenotype was apparent as early as the P3 primordium (Fig. 2I), and cell proliferation activity decreased in P2 and P3 primordia (Fig. 2J, Fig. 3). Although the num ber of cells in the leaf sheaths of sle1 was reduced, the cells were enlarged, keeping the shape of the leaf sheaths less altered (Fig. 5C, D). Plants are able to compensate for a reduction in cell number by an increase in cell size (Horiguchi and Tsukaya, 2011). Therefore, these cellular enlargements in the sle1 leaf sheath may be attributable partly to impaired cell proliferation. Similar compensation was also observed along the length of the leaf blade, in which the reduction in cell numbers was partially compensated by a 20% enlargement of the cell length compared with that of the wild type (Fig. 1H). Along the width of the leaf blade, however, no distinct compensation was observed in the epidermal cells, i.e. the cell width was not altered despite a reduced number of cells across the width of the leaf. Therefore, the degree of compensation may differ depending on the organ and/or the direction, and the phenotypic alteration of sle1 mutants was apparently affected by such a heterogeneous compensation.
The CSLD subfamily genes exhibit the most ancient intron/exon structure, and the wide distribution of CSLD genes across all land plant taxa implies a highly conserved function of this subfamily of proteins (Richmond and Somerville, 2000). In this study, we have presented evidence that OsCSLD4 is required for proper cell division and proliferation during the M phase of the cell cycle. These results reinforce the fundamental function of CSLD genes in plant development. Söllner et al. (2002) concluded that certain aspects of cytokinesis and root-hair morphogenesis might be regulated by the same molecules. Therefore, it is probable that the product of the CSLD subfamily is involved not only in tip growth but also in cell proliferation. In fact, the root-hair defect in the Atcsld3 mutant could be rescued partly by introducing 35S:YFP-AtCSLD5 (Yin et al., 2011). Further study of OsCSLD4 is expected to yield new insights into the role of hemicelluloses in plant development.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Phylogenetic tree of CSLD proteins in rice (Os), maize (Zm), Arabidopsis (At), barley (Hv), sorghum (Sb), and purple false brome (Bradi).
Fig. S2. Comparison of costal cell size in the third-leaf blade of wild-type, sle1-1, and sle1-2 plants.
Fig. S3. Internode elongation patterns in wild-type, sle1-1 and sle1-2 mature plants.
Fig. S4. Root phenotypes of wild-type and sle1 plants.
Fig. S5. Comparison of the number of vascular bundles and the vascular bundle interval in the third-leaf blade among wild-type, sle1-1, and sle1-2 plants.
Fig. S6. Double-target in situ hybridization of OsCLSD4/SLE1 counterstained with histone H4 in a cross-section of shoot apex in wild-type plants.
Fig. S7. Real-time PCR analysis of histone H4, CDKB2;1, and OsCSLD4 expression in wild-type, sle1-1 and sle1-2 plants.
Fig. S8. Expression pattern of AtCSLD5 in a cross-section of Arabidopsis shoot apex.
Acknowledgements
We would like to express great appreciation to Drs Kazuhiko Nishitani and Ryusuke Yokoyama (Tohoku University) for valuable discussions and advice on our work. We also thank Dr Masaaki Umeda (Nara Institute of Science and Technology) for kindly providing the cDNA of CDKB2;1, and Dr Yutaka Sato (Nagoya University) for kind suggestions to help the progress of our work.
Glossary
Abbreviations:
- SE
standard error.
References
- Bernal AJ, Jensen JK, Harholt J, et al. 2007. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis . The Plant Journal 52, 791–802. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WGT. 2008. Functional analysis of the cellulose synthase-like genes Csld1, Csld2, and Csld4 in tip-growing Arabidopsis cells. Plant Physiology 148, 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. 2006. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 311, 1940–1942. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. 2007. A gene from the cellulose synthase-like C family encodes a β-1,4-glucan synthase. Proceedings of the National Academy of Sciences, U S A 104, 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. 2009. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis . Proceedings of the National Academy of Science, USA 106, 5996–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivany FM, Yulia D, Burton RA, Shirley NJ, Wilson SM, Fincher GB, Bacic A, Newbigin E, Doblin MS. 2009. The Cellulose-synthase like C (CslC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Molecular Plant 2, 1025–1039. [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boundonck K, Steer M, Shaw P, Dolan L. 2001. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis . Genes and Development 15, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. 2009. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiology 149, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD. 2002. Cellulose synthase-like genes of rice. Plant Physiology 128, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Tsukaya H. 2011. Organ size regulation in plants: insights from compensation. Frontiers in Plant Science 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhu L, Zeng D, et al. 2010. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Molecular Biology 73, 283–292. [DOI] [PubMed] [Google Scholar]
- Hunter CT, Kirienko DH, Sylvester AW, Peter GF, McCarty DR, Koch KE. 2012. Cellulose synthase-like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiology 158, 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dong B, Shiran B, Talbot MJ, Edlington JE, Hughes T, White RG, Gubler F, Dolferus R. 2011. Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiology 156, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M. 2003. The SCARECROW gene’s role in asymmetric cell divisions in rice plants. The Plant Journal 36, 45–54. [DOI] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han C. 2007. OsCsld1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiology 143, 1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth DL, Williams ND. 2001. Characterization of a mitotic mutant of durum wheat. Chromosome Research 9, 377–386. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. 1993. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Molecular and General Genetics 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. 2002. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proceedings of the National Academy of Sciences, U S A 99, 11109–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. 2009. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. The Plant Journal 60, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. 2005. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proceedings of the National Academy of Sciences, U S A 102, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jügens G. 1996. Cytokinesis in the Arabidopsis embryo involves the synthaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Matsui T, Omasa K, Horie T. 1999. Mechanism of anther dehiscence in rice (Oryza sativa L.). Annals of Botany 84, 501–506. [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH. 2003. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Molecular Biology 53, 423–442. [DOI] [PubMed] [Google Scholar]
- Penning BW, Hunter CT III, Tyengwa R, et al. 2009. Genetic resources for maize cell wall biology. Plant Physiology 151, 1703–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. 2000. The cellulose synthase superfamily. Plant Physiology 124, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH, Jr, Staehelin LA. 1995. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. Journal of Cell Biology 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annual Review of Plant Biology 61, 263–289. [DOI] [PubMed] [Google Scholar]
- Söllner R, Glässer G, Wanner G, Somerville CR, Jürgens G, Assaad FF. 2002. Cytokinesis-defective mutants of Arabidopsis . Plant Physiology 129, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, Kasmi FE, Richter S, Lukowitz W, Assaad FF, Jügens G, Mayer U. 2002. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Current Biology 12, 153–158. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL. 2006. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa . Plant Physiology 142, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG. 2008. Cellulose biosynthesis and deposition in higher plants. New Phytologist 178, 239–252. [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. 1999. Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiology 119, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. 2001. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis . Plant Physiology 126, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Fu Y, Hu G, Si H, Cheng S, Liu W. 2010. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232, 313–324. [DOI] [PubMed] [Google Scholar]
- Yin L, Verhertbruggen Y, Oikawa A, et al. 2011. The cooporative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Molecular Plant 10.1093/mp/ssr026 [DOI] [PubMed] [Google Scholar]
- Zhou S, Wang Y, Li W, et al. 2011. Pollen semi-sterility1 encodes a Kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23, 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH. 2000. KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.