SUMMARY
Eukaryotic ribosome biogenesis requires hundreds of trans-acting factors and dozens of RNAs. Although most factors required for ribosome biogenesis have been identified, little is known about their regulation. Here, we reveal that the yeast deubiquitinating enzyme Ubp10 is localized to the nucleolus and that ubp10Δ cells have reduced pre-rRNAs, mature rRNAs, and translating ribosomes. Through proteomic analyses, we found that Ubp10 interacts with proteins that function in rRNA production and ribosome biogenesis. In particular, we discovered that the largest subunit of RNA polymerase I (RNAPI) is stabilized via Ubp10-mediated deubiquitination and that this is required in order to achieve optimal levels of ribosomes and cell growth. USP36, the human ortholog of Ubp10, complements the ubp10Δ allele for RNAPI stability, pre-rRNA processing, and cell growth in yeast, suggesting that deubiquitination of RNAPI may be conserved in eukaryotes. Our work implicates Ubp10/USP36 as a key regulator of rRNA production through control of RNAPI stability.
INTRODUCTION
Ribosome biogenesis is one of the eukaryotic cell’s most important and energetically costly processes. In budding yeast, cycling cells must make ~2,000 ribosomes per minute, up to 80% of all RNA produced in the cell is rRNA, and 50% of RNA polymerase II transcription events are on ribosomal protein genes (Warner, 1999). Ribosome biogenesis is also an exceptionally complicated process in eukaryotes. It requires hundreds of trans-acting proteins and dozens of RNAs to assemble a functional ribosome (Henras et al., 2008). While global genomic and proteomic studies have led to the identification of the majority of ribosome biogenesis factors (Henras et al., 2008), the exact function for most ribosome biogenesis factors and the regulation that controls their activity still remain to be elucidated.
One of the principal means by which eukaryotic cells regulate protein stability, activity, localization, and/or interactions is through ubiquitination. The covalent attachment of ubiquitin to target proteins is mediated by the action of ubiquitin-protein ligases, and eukaryotic cells express hundreds of ubiquitin-protein ligases that are responsible for the ubiquitination of thousands of different proteins (Ciechanover, 2006). Protein ubiquitination can be reversed by deubiquitinating enzymes (DUBs), which number from 20 in yeast to ~100 in humans (Reyes-Turcu et al., 2009). DUBs provide the cell with the means to modulate the duration of the ubiquitin signal.
Compelling evidence points to a key involvement of ubiquitin in ribosome biogenesis. Ubiquitin itself is initially made as a fusion with ribosomal proteins and this is important for efficient ribosome biogenesis (Finley et al., 1989). In human cells, ubiquitin is highly localized to the nucleolus, multiple complexes involved in ribosome biogenesis are associated with ubiquitin, and a number of nucleolar proteins have increased stability after proteasome inhibition (Chen et al., 2002; Fátyol and Grummt, 2008; Itahana et al., 2003; Stavreva et al., 2006). The human DUB USP36 is nucleolar localized and regulates the stability of the nucleolar proteins nucleophosmin/B23 and fibrillarin (Endo et al., 2009a, 2009b). Despite the centrality of ribosome biogenesis in cellular physiology and the importance of ubiquitin as a posttranslational regulator of protein function, these observations comprise the majority of information available to date about ubiquitin-mediated control in ribosome biogenesis.
Within the catalytic DUB domain, human USP36 shares 49% similarity to budding yeast Ubp10 (Buszczak et al., 2009). To this point, it is not known if yeast Ubp10 localizes to the nucleolus or participates in any aspect of ribosome biogenesis, as is implicated for human USP36 (Endo et al., 2009a, 2009b; Sowa et al., 2009). The only identified role for Ubp10 in yeast is that it deubiquitinates histone H2B to modulate gene silencing at telomere-proximal regions (Emre et al., 2005; Gardner et al., 2005b; Kahana and Gottschling, 1999; Singer et al., 1998). In this report, we conducted a number of genetic, cell biological, and proteomic analyses with Ubp10 and discovered that Ubp10 regulates the ubiquitination and stability of the largest subunit of RNA polymerase I.
RESULTS
Ubp10 Is Necessary for Optimal Cell Growth and Localizes to the Nucleolus
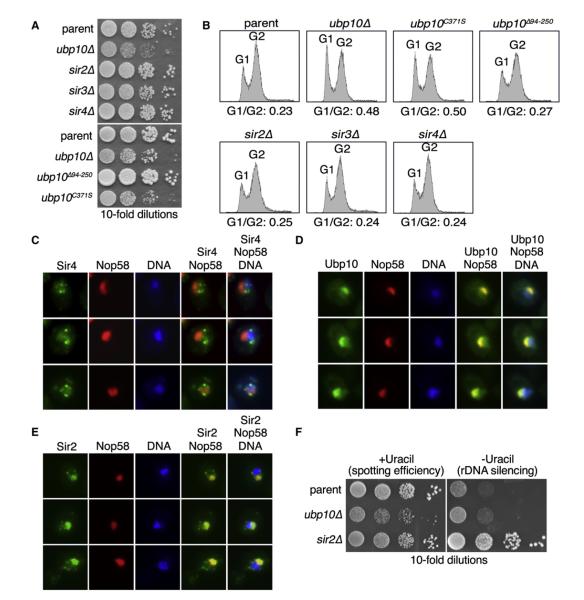
Ubp10’s only known function to date is the deubiquitination of H2B to mediate gene silencing at telomeres (Emre et al., 2005;Gardner et al., 2005b; Kahana and Gottschling, 1999; Singer et al., 1998). However, cells deleted for UBP10 display a slow growth phenotype that cannot be solely explained by loss of telomere gene silencing. As previously noted, ubp10Δ cells have a slow growth phenotype that is correlated with an increased proportion of cells in an asynchronous culture in the G1 phase (Kahana and Gottschling, 1999; Figures 1A and 1B). By contrast, sir2Δ, sir3Δ, and sir4Δ cells, which are deleted for the key chromatin-binding proteins that function in telomere gene silencing (Aparicio et al., 1991), grow identically to parent cells and do not display an increased G1 population (Figures 1A and 1B). In an earlier study, it was found that deletion of residues 94–250 in Ubp10 disrupts Ubp10’s ability to interact with Sir4 (Kahana and Gottschling, 1999), and this mutation specifically ablates Ubp10’s function in telomere gene silencing (Gardner et al., 2005b). As with sir2Δ, sir3Δ, and sir4Δ cells, we found that telomere gene silencing-deficient ubp10Δ94-250 cells grow indistinguishably from parent cells (Figures 1A and 1B). By contrast, mutation of Ubp10’s catalytic residue by creation of a C371S substitution resulted in ubp10C371S cells having an identical slow growth phenotype and proportion of cells in G1 as observed with ubp10Δ cells (Figures 1A and 1B). Thus, optimal cell growth requires intact Ubp10 deubiquitinating enzyme activity, but not intact telomere gene silencing capability.
Figure 1. Ubp10 Is Necessary for Optimal Cell Growth and Localizes to the Nucleolus.
(A) Plate growth of a parent strain compared to ubp10Δ, sir2Δ, sir3Δ, sir4Δ, ubp10Δ94-250, and ubp10C371S strains. Tenfold serial dilutions of cells were spotted on rich medium and incubated at 30°C for 3 days.
(B) Cell-cycle profiles for asynchronous cultures of strains from (A) were examined by propidium iodide staining and flow cytometry. Proportions of cells in G1 versus G2 phases are presented.
(C–E) Fluorescence microscopy of exponentially growing cells coexpressing Nop58-dsRed and either Sir4-GFP (C), Ubp10-GFP (D), or Sir2-GFP (E). Nop58-dsRed marks the nucleolus and DAPI staining marks chromatin.
(F) Spot tests of parent, ubp10Δ, or sir2Δ cells containing the rDNA silencing reporter RDN1::URA3. Cells were spotted in 10-fold serial dilutions onto media containing uracil to assess spotting efficiency or without uracil to measure rDNA silencing. Plates were incubated at 30°C for 3 days.
We examined Ubp10’s cellular localization to see if it differed from that of the Sir4 protein, which interacts with Ubp10 and recruits Ubp10 to telomeres (Gardner et al., 2005b; Kahana and Gottschling, 1999). We found that Sir4 localizes to punctate structures at the edges of the nucleus indicative of telomere clustering as shown previously (Palladino et al., 1993; Figure 1C), whereas Ubp10 is highly enriched in the nucleolus as observed by Ubp10’s colocalization with the nucleolar protein Nop58 (Figure 1D). The predominant nucleolar localization of Ubp10 indicates that Ubp10 likely participates in some aspect of nucleolar biology.
In yeast, gene silencing also occurs at the nucleolar rDNA and this is mediated in part by Sir2 (Bryk et al., 1997; Fritze et al., 1997; Smith and Boeke, 1997), which also has significant nucleolar localization (Gotta et al., 1997; Figure 1E). Because Sir2 has an additional role in rDNA silencing, it is possible that Ubp10 is similarly localized to the nucleolus to mediate rDNA silencing. To assay for a role of Ubp10 in rDNA silencing, we used a previously developed rDNA silencing reporter RDN1::URA3 (Smith and Boeke, 1997), in which the URA3 gene is integrated at the rDNA locus and loss of rDNA silencing is observed by enhanced growth on media that lacks uracil. Using this reporter, rDNA silencing was abolished in sir2Δ cells as previously reported (Smith and Boeke, 1997; Figure 1F), but was intact in ubp10Δ cells (Figure 1F). This agrees with a previous report that loss of UBP10 does not appreciably affect rDNA silencing (Singer et al., 1998).
Loss of Ubp10 Leads to a Reduction 35S Pre-rRNA Levels
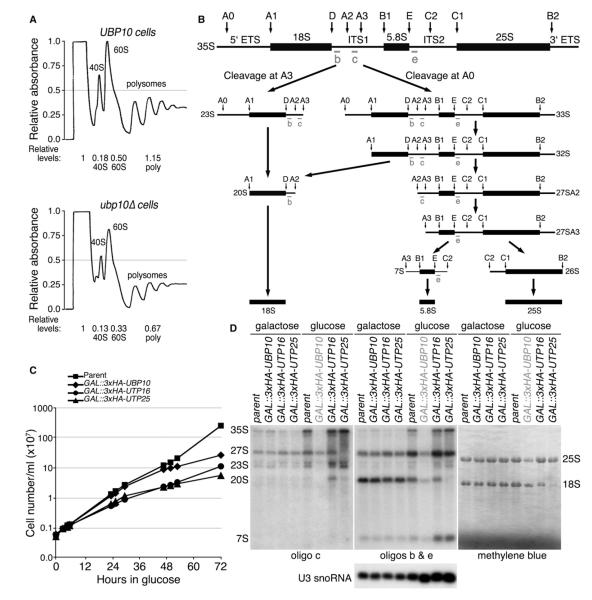
The nucleolus is the site of ribosome biogenesis and it is possible that Ubp10’s function in the nucleolus is to regulate the production of ribosomes. Therefore, we examined polysome profiles generated from cell lysates of UBP10 and ubp10Δ cells to see if ribosome content was altered in ubp10Δ cells. By measuring the area under the curves in the polysomes, we found there was an ~30%–40% reduction in 40S and 60S subunits as well as translating ribosomes in ubp10Δ cells (Figure 2A). The ratio of 40S to 60S subunits was maintained in ubp10Δ cells (0.36 in UBP10 cells versus 0.39 in ubp10Δ cells), suggesting that reduced ribosome content in ubp10Δ cells is due to an overall ribosome production defect. Because ribosomal proteins are known to be ubiquitinated (Spence et al., 2000), one possibility for the reduced ribosome content in ubp10Δ cells is that Ubp10 positively regulates the levels of ribosomal proteins. However, when we examined the levels and stability of select ribosomal proteins from both the small and large ribosomal subunits, we did not observe any decrease in steady-state levels or stability (Figure S1).
Figure 2. Ubp10 Is Required for Optimal Ribosome Biogenesis.
(A) Polysome profiles for UBP10 and ubp10Δ cells were generated from lysates of asynchronous cultures. Lysates were loaded onto 7%–47% sucrose gradients and centrifuged for 1.5 hr at 39,000 rpm at 4°C. Preps were normalized to A260 units. The top of the gradient is to the left.
(B) Schematic of the pre-rRNA processing pathway in yeast.
(C) Genetic depletion of Ubp10 resulted in a slow growth phenotype. Number of cells per ml are plotted versus time in glucose growth medium at 17°C. The parent strain (YPH499), the GAL1::3xHA-UBP10 strain, and two control strains1 (GAL1::3xHA-UTP16 and GAL1::3xHA-UTP25) are shown.
(D) Genetic depletion of Ubp10 resulted in a decrease in pre-rRNA species and mature 18S and 25S rRNA. Total RNA was extracted from yeast in which the indicated protein was genetically depleted for 72 hr at 17°C. Mature and pre-rRNA species were detected by northern analysis using the indicated oligonucleotide probes or with methylene blue staining. U3 snoRNA serves as a control. The parental strain (YPH499) is shown along with GAL1::3xHA-UTP16 and GAL1::3xHA-UTP25 as controls.
See also Figure S1.
We performed a northern analysis to examine if Ubp10 plays a role in the processing steps required to cleave the 35S pre-rRNA transcript into the mature 18S, 5.8S, and 25S rRNAs (Figure 2B). For these studies, we placed the GAL1 promoter in front of the endogenous UBP10 gene, thus creating a GAL1::3HA-UBP10 strain in which Ubp10 can be conditionally depleted upon transfer of cells to glucose growth medium. As expected from the slow growth phenotype of ubp10Δ cells (Figure 1A), genetic depletion of Ubp10 in GAL1::3HA-UBP10 cells resulted in a slow growth phenotype that was observable 48 hr after the switch to glucose growth medium (Figure 2C). Genetic depletion of Ubp10 resulted in a decrease in the abundance of several rRNA precursor species, but most notably the 35S pre-rRNA (Figure 2D). Consistent with the reduction of 40S and 60S subunits in the ubp10Δ polysome profiles, we also observed a ~40% decrease in the levels of the mature 18S and 25S rRNAs after genetic depletion of Ubp10. By contrast, genetic depletion of Utp16 and Utp25—two factors involved in 18S rRNA processing (Charette and Baserga, 2010; Dragon et al., 2002)—led to accumulation of the 35S primary transcript but depletion of the mature 18S rRNA (Figure 2D). Overall, loss of Ubp10 resulted in a defect in 35S pre-rRNA levels that in turn likely affects production of the large and small subunit rRNAs and reduces total mature ribosome levels in the cell.
Proteins that Interact with Ubp10 Are Enriched in Ribosome Biogenesis Functions
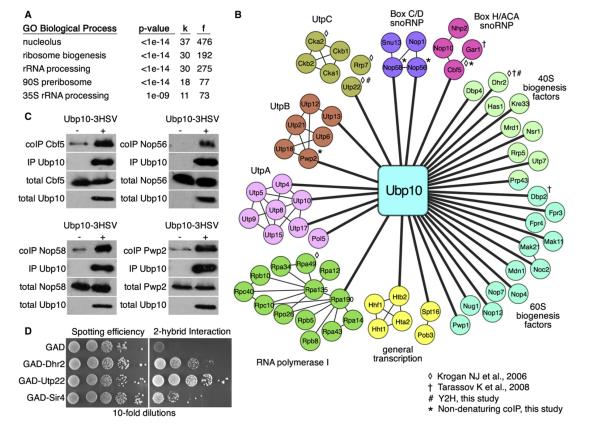
The nucleolar localization of Ubp10 and reduced 35S pre-rRNA and mature ribosome content in Ubp10-deficient cells indicated that Ubp10 likely interacts with nucleolar proteins and has at least one nucleolar ribosome biogenesis factor as a substrate. To identify Ubp10-interacting proteins and substrates, we used a coimmunoprecipitation (coIP) tandem mass spectrometry (MS/MS) approach. Because enzyme-substrate interactions are often transient, we used in vivo formaldehyde crosslinking to maintain both stable and transient Ubp10 interactions during the coIP procedure (Sutherland et al., 2008). This strategy also allowed us to lyse cells in strong denaturing conditions (8 M urea, 1% SDS) to minimize nonspecific interactions. We avoided excessive crosslinking by using a duration of crosslinking that caused 50% of the total Ubp10 cellular pool to incorporate into high molecular weight crosslinked species as observed by western analysis. To facilitate the coIP, we used a C-terminal 3HSV-tagged version of Ubp10 expressed from its native promoter. Addition of the 3HSV tag did not affect Ubp10’s functions in cell growth, telomere gene silencing, or histone H2B deubiquitination (Figure S2). We used untagged Ubp10 as a control. All coIPs were performed in triplicate with independently grown cultures. We categorized a protein as interacting with Ubp10 if the summed spectral (peptide) counts of the protein identified by MS/MS in the tagged replicates was a minimum of 4 and exceeded its summed spectral counts in the untagged replicates by ≥3-fold. Using these criteria, we identified 83 Ubp10-interacting proteins (Table S1; Ubp10 PP filter ≥0.35).
We analyzed the set of Ubp10 interactors using the cluster interpreter FunSpec (http://funspec.med.utoronto.ca/), which can reveal enrichment for cellular functions, localizations, known protein complexes, and other useful parameters (Robinson et al., 2002). Of the 83 candidates, 37 have full or partial nucleolar localization (Figure 3A). The interactors are also enriched for proteins involved in ribosome biogenesis and, more specifically, in 35S pre-rRNA processing as part of the 90S preribosome (Figure 3A). It is possible that ribosome biogenesis proteins are present in the Ubp10-interaction data set because they nonspecifically coIP as a general result of the procedure. However, we also performed similar crosslinking coIP MS/MS analyses with two additional DUBs (Ubp13 and Otu1) and did not observe any enrichment for ribosome biogenesis factors (Table S1).
Figure 3. Ubp10 Interacts with Ribosome Biogenesis Proteins.
(A) FunSpec (http://funspec.med.utoronto.ca/) analysis of the 83 interacting proteins identified by crosslinking coIP MS/MS with Ubp10-3HSV. The p value represents the probability that the intersection of a given list with any functional category occurs by chance. “k” represents the number of genes from the input and “f” represents the total number of genes for that category in the yeast proteome.
(B) Network diagram of the nucleolar-localized proteins identified by crosslinking coIP MS/MS with Ubp10-3HSV. Diagram was generated using Cytoscape (http://www.cytoscape.org/) (Smoot et al., 2011). Thick lines connect interactions discovered by the crosslinking coIP MS/MS analysis. Thin lines mark interactions previously described. RNA polymerase I, BoxC/D snoRNP, and Box H/ACA snoRNP interactions were derived from solved crystal structures (Kuhn et al., 2007; Liang et al., 2009; Rashid et al., 2006; Reichow et al., 2007). UtpA, UtpB, and UtpC interactions were derived from global coIP and Y2H studies (Dragon et al., 2002; Gallagher et al., 2004; Grandi et al., 2002; Krogan et al., 2004; Kuhn et al., 2007; Li and Ye, 2006; Liang et al., 2009; Lim et al., 2011; Rashid et al., 2006;Reichow et al., 2007; Tarassov et al., 2008; Wittmeyer et al., 1999). ◇ and † denote those interactions between Ubp10 and the target proteins that have been previously published (Krogan et al., 2006; Tarassov et al., 2008). # and * denote those interactions between Ubp10 and the target proteins that also occur by coIP in (C) or Y2H in (D).
(C) coIPs between Ubp10-3HSV and the indicated 3HA-tagged target proteins. Ubp10-3HSV was purified with its interacting proteins by immunoprecipitation using anti-HSV antibodies. 1% of lysates and 10% of coIPs were separated by SDS-PAGE and analyzed by anti-HSV and anti-HA western blotting.
(D) Yeast 2-hybrid (Y2H) interactions between cells expressing GBD-Ubp10 and each indicated GAD fusion protein. Cells were spotted onto media plus or minus histidine to measure spotting efficiency and Y2H interaction, respectively.
See also Figure S2.
To aid in visualizing the MS/MS interaction data, we created a network of the nucleolar proteins that interact with Ubp10 (Figure 3B). The thick lines in the network that connect Ubp10 to each protein indicate the primary interactions identified by crosslinking coIP MS/MS. The thin lines represent interactions indicated by previous studies (Dragon et al., 2002; Gallagher et al., 2004; Grandi et al., 2002; Krogan et al., 2004; Kuhn et al., 2007; Li and Ye, 2006; Liang et al., 2009; Lim et al., 2011; Rashid et al., 2006; Reichow et al., 2007; Tarassov et al., 2008; Wittmeyer et al., 1999). From this arrangement, Ubp10 showed interactions with numerous subcomplexes involved in 35S pre-rRNA transcription and processing. In particular, Ubp10 showed an interaction with Rpa190, the essential largest subunit of RNA polymerase I (Mémet et al., 1988), which is responsible for transcribing the 35S pre-rRNA (Schneider, 2012). Ubp10 also showed interactions with proteins of the UtpA (also known as t-Utp), UtpB, and UtpC complexes, which are required for the cleavage events that liberate the 18S rRNA from the 35S pre-rRNA (Phipps et al., 2011). Ubp10 showed interactions with components of the Box C/D and Box H/ACA small nucleolar ribonucleoprotein particles (snoRNPs), which are involved in rRNA cleavage and modification (Reichow et al., 2007). Ubp10 also showed interactions with several other proteins required for small or large ribosome subunit biogenesis, including a number of key RNA helicases (Dbp2, Dbp4, Dhr2, Has1, and Prp43). Last, we identified histone H2B (Htb2) as a Ubp10-interacting protein, indicating that the method allowed for the capture of Ubp10’s only known substrate.
A main limitation of the crosslinking coIP analysis is that it cannot distinguish between stable and transient interactions. Therefore, to gain insight into how many interactions might be due to stable association with Ubp10, we chose 15 of the 37 nucleolar proteins (Cbf5, Dhr2, Htb2, Kre33, Noc2, Nsr1, Nop1, Nop56, Nop58, Pwp2, Rpa190, Rrp9, Rrp17, Utp13, and Utp22) to test for a stable interaction with Ubp10 by traditional coIP and also by yeast 2-hybrid (Y2H) assay. To facilitate the coIP, we added a 3HA tag to the C terminus of the Ubp10-interacting proteins in a strain expressing Ubp10-3HSV (in the case of Htb2, we used a previously constructed FLAG-tagged version; Gardner et al., 2005b). For the Y2H, we tested if a Gal4 binding domain (GBD) fusion to Ubp10 could interact with Gal4 activation domain (GAD) fusions of the target proteins. Of the 15, 6 retested as positive interactors, 4 showed an interaction by traditional by coIP (Cbf5, Nop56, Nop58, and Pwp2), and 2 showed an interaction by Y2H (Dhr2 and Utp22) (Figures 3C and 3D). Of the four proteins that interacted with Ubp10 by traditional coIP, Pwp2 showed an interaction when the reciprocal coIP was performed (Figure S2C). Altogether, some of the interacting proteins identified by crosslinking coIP MS/MS form stable interactions with Ubp10 that can be queried by traditional coIP.
The Ubp10 protein interactions identified by crosslinking coIP that did not confirm by traditional coIP potentially represent proteins that transiently interact with Ubp10, and thus can only be captured under crosslinking conditions. Consistent with this, we identified Ubp10’s known substrate histone H2B (Htb2) in the crosslinking coIP MS/MS analysis, but we were unable to establish an interaction between Ubp10 and histone H2B by coIP. However, it is important to note that nonspecific interactions will also fail to verify by traditional coIP, and thus the list of Ubp10 interactors identified here by crosslinking coIP could also contain some nonspecific interactions. Paring the final list to only bona fide Ubp10-interacting proteins will require extensive additional studies for each putative interacting protein. In the following sections, we present our studies focusing on one of these: Rpa190.
Ubp10 Regulates the Ubiquitination and Stability of the Largest Subunit of RNA Polymerase I
We examined the list of Ubp10 interactors for a protein that, if it were a substrate of Ubp10, could explain the reduction of 35S pre-rRNA and mature ribosome content in Ubp10-deficient cells. The most conspicuous was Rpa190, the largest subunit of RNA polymerase I (RNAPI) (Mémet et al., 1988), which is responsible for transcription of the 35S pre-rRNA (Schneider, 2012). Dysregulation of Rpa190 and rDNA transcription in ubp10Δ cells would provide the most parsimonious explanation for the reduced 35S pre-rRNA and mature 18S and 25S rRNA levels.
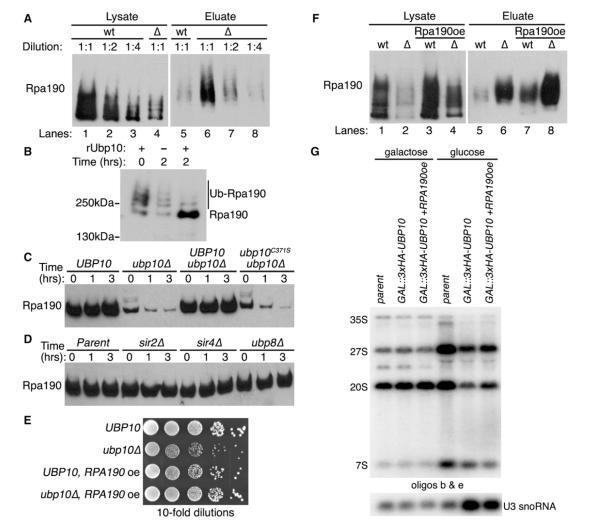
Similar to histone H2B, we did not observe an interaction between Ubp10 and Rpa190 by traditional coIP, suggesting that the Ubp10 and Rpa190 might only transiently associate via an enzyme-substrate interaction. Therefore, we determined if Rpa190 was a substrate of Ubp10 by examining if Rpa190 is ubiquitinated and if Ubp10 regulates that ubiquitination. To do this, we added a 3HA tag to the C terminus of Rpa190 expressed from its genomic locus in cells that also express ubiquitin with an 8His tag at its N terminus. We prepared lysates from UBP10 or ubp10Δ cells under heavily denaturing conditions and purified their respective ubiquitin proteomes by metal affinity chromatography. We then examined both the initial cell lysates for steady-state Rpa190 protein levels and the eluted ubiquitin proteomes for steady-state Rpa190 ubiquitination levels. We found that Rpa190 steady-state protein levels were ~5-fold lower in ubp10Δ cells than in UBP10 cells (Figure 4A, left panel), suggesting that Ubp10 might control Rpa190 stability. Despite the lower protein levels of Rpa190 in ubp10Δ cells, we found that the steady-state ubiquitination levels of Rpa190 were ~4-fold higher in ubp10Δ cells (Figure 4A, right panel). This is consistent with ubiquitinated Rpa190 serving as an in vivo substrate for Ubp10. To determine if Ubp10 is capable of directly deubiquitinating Rpa190 in vitro, we isolated ubiquitinated Rpa190 from ubp10Δ cells and added recombinant Ubp10 purified from Escherichia coli. Addition of Ubp10 resulted in the deubiquitination of Rpa190 (Figure 4B). Thus, Rpa190 appears to be a substrate of Ubp10 both in vivo and in vitro.
Figure 4. Ubp10 Controls Rpa190 Ubiquitination and Stability.
(A) Ubiquitin proteomes from UBP10 (wt) or ubp10Δ (Δ) cells were isolated by metal affinity purification. Levels of Rpa190-3HA in lysates (total protein) and eluates (ubiquitinated Rpa190) were determined by western analysis using anti-HA antibodies. Lanes 1 and 4 indicate steady-state levels of Rpa190 in total lysates of UBP10 and ubp10Δ cells. Lanes 5 and 6 show levels of ubiquitinated Rpa190 in the ubiquitin proteome of UBP10 and ubp10Δ cells.
(B) In vitro Rpa190 deubiquitination assay. Recombinant Ubp10 was added to purified ubiquitinated Rpa190 for the indicated time. Western analysis of Rpa190 was performed using anti-HA antibodies.
(C) Cycloheximide-chase degradation assays of UBP10, ubp10Δ, UBP10-3HSV ubp10Δ, or ubp10C371S-3HSV ubp10Δ cells expressing Rpa190-3HA. Time after cycloheximide addition is indicated above each lane. Western analysis of whole-cell extracts was performed using anti-HA antibodies.
(D) Cycloheximide-chase degradation assays of UBP10, sir2Δ, sir4Δ, or ubp8Δ cells expressing Rpa190-3HA. Assay performed as in (C).
(E) Growth of UBP10 and ubp10Δ cells with or without RPA190 overexpression (oe). Tenfold serial dilutions of cells were spotted onto the appropriate media and incubated at 30°C for 3 days.
(F) Ubiquitin proteome analysis from UBP10 (wt) or ubp10Δ (Δ) cells overexpressing (oe) Rpa190 was performed as in (A).
(G) RPA190 overexpression partially rescues the decrease in pre-rRNA species when UBP10 is genetically depleted. Total RNA was extracted from yeast in which the indicated protein was genetically depleted for 72 hr at 17°C. Pre-rRNA species were detected by northern analysis using the indicated oligonucleotide probes from Figure 2B.
We observed lower steady-state protein levels of Rpa190 in ubp10Δ cells (Figure 4A), and this could be the result of either decreased Rpa190 protein stability due to increased ubiquitination or decreased RPA190 gene transcription. We previously conducted a transcript microarray analysis to determine which genes had altered transcription in ubp10Δ cells (Gardner et al., 2005b). From that analysis, we found RPA190 transcription is increased by approximately 50% in ubp10Δ cells, indicating that reduced transcription is not the explanation for the low levels of Rpa190. Therefore, we assessed Rpa190 protein stability in either the presence or absence of UBP10 using a standard cycloheximide-chase degradation assay. We found that Rpa190 was stable in UBP10 cells whereas Rpa190 had a significantly lower steady-state level and was degraded in ubp10Δ cells (Figure 4C). This effect is directly due to loss of UBP10 and not an additional mutation in the ubp10Δ cells because the stability of Rpa190 was rescued by addition of UBP10 (Figure 4C). By contrast, addition of catalytically inactive ubp10C371S did not rescue the stability of Rpa190 indicating that Ubp10’s DUB activity is required to stabilize Rpa190 (Figure 4C). We did not observe any alterations in Rpa190 stability in sir2Δ, sir4Δ, and ubp8Δ cells (Figure 4D), indicating that control of Rpa190 stability is not related to loss of rDNA silencing (sir2Δ), telomere gene silencing (sir4Δ), or general histone H2B deubiquitination (ubp8Δ).
A drastic reduction in Rpa190 levels could explain the slow growth phenotype of ubp10Δ cells. To test this idea, we overexpressed RPA190 from a high copy plasmid. We found that increased expression of Rpa190 rescued the slow growth phenotype of ubp10Δ cells (Figure 4E). Under the overexpression conditions tested, Rpa190 levels in ubp10Δ cells with overexpression of RPA190 were ~50% of Rpa190 levels in UBP10 cells with normal RPA190 expression (Figure 4F, compare lanes 1 and 4). Moreover, we found that overexpression of RPA190 partially rescued the deficit in pre-rRNA species’ levels in cells genetically depleted for UBP10 (Figure 4G). The partial rescue of pre-rRNA species coupled with the full rescue of growth suggests that optimal growth does not require maximal levels of Rpa190 or maximum production of rRNA. However, it does indicate that the loss of Ubp10’s control over Rpa190 levels is sufficient to explain the growth defect of UBP10-deficient cells.
Rpa190 Degradation Is Not Nutrient or DNA-Damage Sensitive, but Is Cold Sensitive
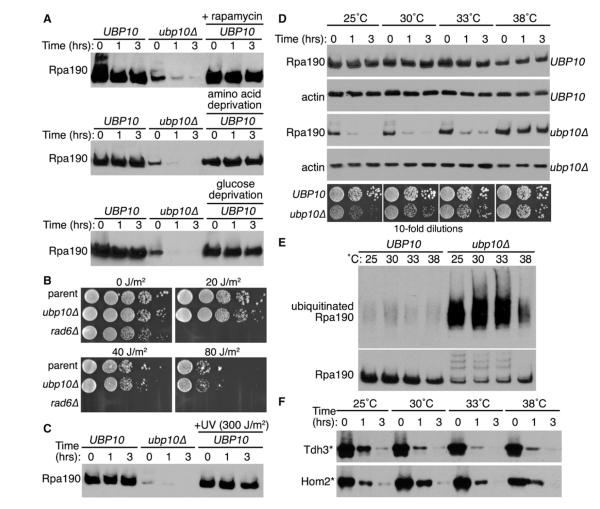
One of the key conditions known to regulate rDNA transcription is nutrient availability (Lempiäinen and Shore, 2009): scarce nutrient availability decreases rDNA transcription, whereas abundant nutrient availability increases rDNA transcription. Accordingly, the most obvious hypothesis for Ubp10-dependent regulation of Rpa190 stability is that Ubp10 functions to remove ubiquitin from Rpa190 to prevent its degradation when nutrients are abundantly available, thereby maximizing the amount of RNAPI that can function in rDNA transcription and ribosome biogenesis. The prediction from this hypothesis is that, under limiting nutrient availability, Ubp10 would not deubiquitinate Rpa190 and this would result in the degradation of Rpa190 in UBP10 cells.
When we initially searched the literature, we found that most studies pertaining to nutrient regulation of rRNA transcription have focused on the transcriptional regulation of ribosomal protein genes or the rDNA itself (Lempiäinen and Shore, 2009). Few studies have explored the effects of nutrient regulation at the level of ribosome biogenesis factor protein stability. However, it was recently discovered that the steady-state levels of Rrn3, a transcription factor that recruits the RNAPI holoenzyme to the rDNA promoter (Yamamoto et al., 1996), are dramatically reduced after target of rapamycin inhibition by rapamycin or amino acid depletion from the growth media, and this is due to proteasome-dependent degradation of Rrn3 (Philippi et al., 2010). By contrast, it was demonstrated in an earlier study that Rpa190 steady-state levels are unaffected after rapamycin treatment (Tsang et al., 2003), suggesting that Rpa190 stability is not nutrient sensitive in UBP10 cells. To test this directly, we examined Rpa190 stability after a similar rapamycin treatment (200 nM), amino acid (tryptophan) depletion, or glucose depletion. In no case did we observe any effect on Rpa190 stability in UBP10 cells after these manipulations (Figure 5A), demonstrating that Rpa190 stability is not regulated by nutrient availability.
Figure 5. Rpa190 Degradation Is Not Nutrient or DNA-Damage Sensitive, but Is Cold Sensitive.
(A) Cycloheximide-chase degradation assays of UBP10 or ubp10Δ cells expressing Rpa190-3HA. Cells were subject to rapamycin treatment (200 nM), amino acid depletion (media with no tryptophan), or glucose depletion (media with no glucose) for 2 hr prior to addition of cycloheximide. Time after cycloheximide addition is indicated above each lane. Western analysis was performed using anti-HA antibodies.
(B) Growth of UBP10 and ubp10Δ cells (left panels) with or without UV exposure. Tenfold serial dilutions of cells were spotted onto the appropriate media and exposed to the indicated amount of UV irradiation. rad6Δ cells were used as a positive control. Plates were subsequently incubated at 30°C for 3 days.
(C) Cycloheximide-chase degradation assays of UBP10 or ubp10Δ cells expressing Rpa190-3HA after UV exposure. Cells were exposed to the indicated amount of UV irradiation, and cycloheximide was subsequently added. Time after cycloheximide addition is indicated above each lane. Western analysis was performed using anti-HA antibodies.
(D) Cycloheximide-chase degradation assays of UBP10 or ubp10Δ cells expressing Rpa190-3HA. Cells were grown at the indicated temperature. Time after cycloheximide addition is indicated above each lane. Western analysis was performed using anti-HA antibodies. Growth of UBP10 and ubp10Δ cells (bottom panels). Tenfold serial dilutions of cells were spotted onto the appropriate media and incubated at the indicated temperature for 3 days (30°C, 33°C, or 38°C) or 5 days (25°C).
(E) Ubiquitin proteomes from UBP10 (wt) or ubp10Δ (Δ) cells incubated at the indicated temperatures were isolated by metal affinity purification. Levels of Rpa190-3HA in lysates (bottom panel) and eluates (top panel) were determined by western analysis using anti-HA antibodies.
(F) Cycloheximide-chase degradation assays of cells expressing two San1 substrates (Tdh3* and Hom2*) were performed as in (B).
In yeast, the DUB Ubp3 regulates the stability of Rpb1, the largest subunit of RNA polymerase II (RNAPII), in response to UV-induced DNA damage (Kvint et al., 2008). Therefore, we examined if Ubp10 might have an analogous function in controlling Rpa190 stability after exposure to UV. We found no sensitivity of ubp10Δ cells to increasing doses of UV exposure (Figure 5B). Furthermore, the stability of Rpa190 was unchanged in UBP10 cells after exposure to UV (Figure 5C). From this, we conclude that Ubp10 deubiquitination of Rpa190 serves a different purpose than Ubp3 deubiquitination of Rpb1.
Interested in what conditions might regulate Rpa190 stability, we examined a number of other growth and stress conditions to determine their affect on Rpa190 degradation in ubp10Δ cells. We found that Rpa190 degradation in ubp10Δ cells occurred as a function of the growth temperature. As the growth temperature was increased, the stability of Rpa190 in ubp10Δ cells also increased (Figure 5D), and Rpa190 ubiquitination in ubp10Δ cells decreased (Figure 5E). The increased stability of Rpa190 in ubp10Δ cells at higher temperatures was not due to a general reduction in ubiquitin-mediated proteasome degradation as we observed little change in the proteasome degradation of nuclear proteins ubiquitinated by the nuclear ubiquitin ligase San1 (Figure 5F; Rosenbaum et al., 2011). Increased Rpa190 stability in ubp10Δ cells also correlated with the observation that the slow growth phenotype of ubp10Δ cells was progressively ameliorated when the cells were grown at increasingly higher temperatures (Figure 5D). We speculate that ubiquitination of Rpa190 serves as a checkpoint for a cold-sensitive step during rRNA transcription and Ubp10’s function is to remove ubiquitin from Rpa190 thereby stabilizing Rpa190 once that checkpoint has been resolved (see Discussion).
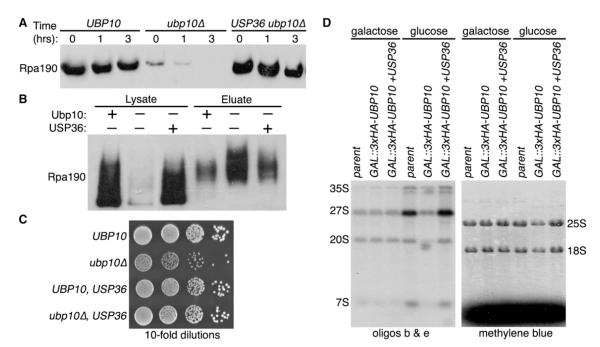
Human USP36 Is a Functional Analog of Ubp10 for RNAPI Ubiquitination and Stability
The human DUB USP36 and the Drosophila DUB scrawny have significant homology with Ubp10 (Buszczak et al., 2009). USP36 shares 27% identity and 49% similarity with Ubp10 in the catalytic DUB domain, and scrawny shares 31% identity and 50% similarity. Interestingly, both USP36 and scrawny are predominantly nucleolar localized in human and Drosophila cells, respectively (Buszczak et al., 2009; Endo et al., 2009b). Similar to the growth deficit, we observed with ubp10Δ yeast cells, knockdown of USP36 levels reduced proliferation of HeLa cells (Endo et al., 2009b), and deletion of scrawny reduced the proliferation of follicle and intestinal stem cells in Drosophila (Buszczak et al., 2009). These observations suggest that human USP36 and Drosophila scrawny might function analogously in modulating RNAPI stability.
Because ribosome biogenesis is fundamental to all organisms, we wondered if a metazoan DUB ortholog could functionally complement the ubp10Δ allele in yeast for Rpa190 ubiquitination and stability. To test this, we placed human USP36 behind the UBP10 promoter in a yeast expression plasmid and transformed the plasmid into ubp10Δ cells. We found that expression of human USP36 restored both Rpa190 stability and ubiquitination levels to those observed in UBP10 cells (Figures 6A and 6B). Furthermore, expression of USP36 rescued the slow growth phenotype observed with ubp10Δ cells (Figure 6C). Lastly, USP36 expression rescued the reduced levels of pre-rRNA species and mature 18S and 25S rRNAs observed after genetic depletion of UBP10 (Figure 6D). Thus, Ubp10/USP36 regulation of RNAPI stability is likely to be a conserved feature of eukaryotic ribosome biogenesis.
Figure 6. USP36 Is a Functional Analog of Ubp10 for RNAPI Ubiquitination and Stability.
(A) Cycloheximide-chase degradation assays of UBP10, ubp10Δ, or USP36 ubp10Δ cells expressing Rpa190-3HA. Time after cycloheximide addition is indicated above each lane. Western analysis of whole-cell extracts was performed using anti-HA antibodies.
(B) Ubiquitin proteomes from UBP10, ubp10Δ, or USP36 ubp10Δ cells were isolated by metal affinity purification. Levels of Rpa190-3HA in lysates (total protein) and eluates (ubiquitinated Rpa190) were determined by western analysis using anti-HA antibodies.
(C) Growth of UBP10, ubp10Δ, or USP36 ubp10Δ strains. Tenfold serial dilutions of cells were spotted onto the appropriate media and incubated at 30°C for 3 days.
(D) rRNA processing analysis of GAL::3xHA-UBP10 cells with or without USP36 overexpression. Total RNA was extracted and analyzed as in Figure 2D.
DISCUSSION
Prior to our studies here, the only known role for Ubp10 was its regulation of gene silencing at the telomeres via deubiquitination of histone H2B (Emre et al., 2005; Gardner et al., 2005b). We have now discovered an additional function for Ubp10 in regulating ribosome biogenesis by control of RNAPI ubiquitination and stability.
Possibilities for RNA Polymerase I Ubiquitin-Mediated Regulation
There is only one other example where deubiquitination of the largest subunit of an RNA polymerase functions to control its stability. In budding yeast, the DUB Ubp3 regulates the ubiquitination and stability of Rpb1 (Kvint et al., 2008), the largest subunit of RNAPII. Cells deleted for UBP3 are sensitive to the transcription elongation inhibitor 6-azauracil (Kvint et al., 2008), suggesting that Rpb1 ubiquitination and Ubp3-dependent deubiquitination play a general role in the rescue of elongation-arrested RNAPII complexes. Consistent with this, ubiquitination of Rpb1 is particularly pronounced in ubp3Δ cells under DNA damage conditions that arrest RNAPII complexes (Kvint et al., 2008). It has been proposed that Ubp3 surveys the ubiquitination status of RNAPII to prevent unwarranted destruction of an arrested RNAPII complex if the situation can be resolved (Kvint et al., 2008).
Although it appears that regulation of Rpa190 stability in UBP10 cells is not controlled by DNA damage or nutrient availability, there are other possibilities for ubiquitin-mediated modulation of RNAPI function. For example, it could be that Rpa190 is ubiquitinated and deubiquitinated to regulate a particular step in rDNA transcription such as initiation, elongation, termination, or reinitiation of transcription. Of particular interest in terms of Ubp10 regulation is the temperature dependence of RNAPI degradation in ubp10Δ cells. Ribosome assembly is inherently sensitive to low temperatures (Guthrie et al., 1969). It is possible that the assembling rRNA transcript has an increasing probability of becoming kinetically trapped in an unproductive folding intermediate at lower temperatures (Treiber and Williamson, 2001), which might lead to the arrest of the elongating RNAPI complex. Ubiquitination of the arrested RNAPI complex could serve as a timing mechanism for resolution of the rRNA kinetic trap. Ubp10’s function in this regard would be to deubiquitinate RNAPI complexes once the trap has been resolved, allowing the resolved RNAPI complex to proceed with transcription and ribosome assembly. At higher temperatures, kinetically trapped rRNA transcripts might be more easily resolved (or more easily avoided) due to increased free energy, and thus there would be fewer arrested RNAPI complexes requiring ubiquitination and deubiquitination. Future work will be required to delineate where, when, and for what purpose Ubp10 regulates RNAPI stability.
Other Nucleolar Functions for Ubp10
Although we highlighted a functional role for Ubp10 in stabilizing RNAPI, it is possible that Ubp10 functions in other aspects of ribosome biogenesis based on the results from our proteomic studies. Interestingly, previously conducted high-throughput proteomic analyses of DUBs in human cells (Sowa et al., 2009) and fission yeast (Kouranti et al., 2010) found that Ubp10’s orthologs (USP36 in humans and Ubp16 in fission yeast) interact with some of the same ribosome biogenesis factors that we identified in our study. For example, USP36 was found to interact with the UtpB complex and DHX33/Prp43 (Sowa et al., 2009), and Ubp16 was found to interact with Prh1/Dhr2, Gar2/Nsr1, and Nop58 (Kouranti et al., 2010). These observations strengthen the idea that Ubp10 and its orthologs might have broader functions in ribosome biogenesis. While the human and fission yeast studies relied on traditional affinity purification methods that primarily query stable protein interactions, our studies used crosslinking to attempt the capture of both stable and transient interactions, thus providing the opportunity to discover a wider spectrum of interactions for Ubp10. It would be intriguing to see if analogous crosslinking studies in human cells and fission yeast would similarly expand the repertoire of Ubp10 ortholog interactions.
Regulation of RNA Polymerase I Stability and Cancer
Ubp10’s human ortholog USP36 shows increased expression in ovarian cancer cells (Li et al., 2008). In light of the fact that Ubp10 is important for cell growth in yeast, USP36 is important for cell proliferation in vitro (Endo et al., 2009b), and the Drosophila homolog scrawny is important for stem cell proliferation in vivo (Buszczak et al., 2009), it is conceivable that cells with high proliferative capacity like cancer cells require increased levels of USP36 to achieve the elevated levels of RNAPI and ribosome production required for rapid proliferation. This possibility is especially salient when framed against the growing body of literature implicating upregulation of ribosome biogenesis as a key factor in tumorigenesis (Montanaro et al., 2008). More studies will be needed to assess if USP36 has true oncogenic potential due its regulation of RNAPI stability and/or some other critical step in ribosome biogenesis.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast strains and plasmids are listed in Table S2. Standard yeast genetic methods were used in these studies (Guthrie and Fink, 1991).
Fluorescence Microscopy
Cells were grown in 5 ml cultures to a density of ~1.8 × 107 cells/ml. Harvested cells were fixed with 4% paraformaldehyde in 0.1 M sorbitol for 15 min, washed once with wash buffer (1.2 M sorbitol, 0.4 M KPO4), stained with DAPI for 5 min in wash buffer plus 2% Triton X-100 and washed two times in wash buffer. Cells were imaged on a Nikon Eclipse 90i with a 100× objective (DIC N2 N.A. 1.4), filters for green (ET470/40×, T495LP, ET525/50 m) or red (ET560/40×, T585LP, ET630/75 m) fluorescence, and a Photometrics Cool Snap HQ2 cooled CCD camera with NIS-Elements acquisition software. All images were processed using Photoshop CS (Adobe Systems Inc.).
Cell-Cycle Analyses
Yeast cell-cycle analyses were conducted similar to those previously described (Haase and Reed, 2002). Cells were grown in 5 ml cultures to a density of ~0.8 × 107 cells/ml, harvested by centrifugation, fixed in 70% ethanol, washed in 50 mM sodium citrate containing 0.25 mg/ml RNase A, incubated at 95°C for 15 min and then 37°C for 2 hr. Fixed cells were resuspended in 50 mM sodium citrate, stained with 2 μM SYTOX green, and sonicated for 1 s before analysis by flow cytometry. G1 and G2 phase analyses was done with WinCycle Software.
Formaldehyde Crosslinking Coimmunoprecipitation
Crosslinking experiments were conducted similar to those previously described (Rosenbaum et al., 2011). Cells were grown in 500 ml cultures to a density of ~1.8 × 107 cells/ml. Formaldehyde was added to a final concentration of 1% (v/v), and cultures incubated for 5 min at 30°C. Crosslinking was quenched with glycine added to a final concentration of 125 mM. Harvested cells were lysed in SUME buffer (8 M Urea, 1% SDS, 10 mM MOPS [pH 6.8], 10 mM EDTA) and lysates were diluted 1:5 in IP buffer (15 mM Na2HPO4, 150 mM NaCl, 2% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 10 mM EDTA). Lysates were incubated with 1:1,000 mouse anti-HA antibody (Novagen) preconjugated to 1.25 mg/ml Protein A Dynabeads (Invitrogen) for 16 hr at 4°C. Beads were washed three times with IP buffer and proteins were eluted by incubation at 65°C for 10 min in 50 μl SUMEB (SUME + 0.01% bromophenol blue).
Mass Spectrometry and Data Analysis
Samples analyzed by MS/MS were run 1 cm into an 8%–16% SDS-PAGE gel and gel slices excised. Proteins in the gel slices were digested with trypsin (see Extended Experimental Procedures). The digestion products were desalted and dried by vacuum centrifugation. Dried peptide mixtures were resuspended in 7 μl of 0.1% formic acid and 5 μl was analyzed by LC/ESI MS/MS using either an LTQ-FT or LTQ-OrbiTrap mass spectrometer (ThermoElectron). Complete MS/MS methods are in the Extended Experimental Procedures. The protein database search algorithm X!Tandem (Craig and Beavis, 2004) was used to identify peptides from the Saccharomyces Genome Database (http://www.yeastgenome.org). Peptide false discovery rates were measured using Peptide Prophet (Keller et al., 2002), and results were stored and analyzed in the Computational Proteomics Analysis System (Rauch et al., 2006). To apply different levels of stringency, peptides were filtered using Peptide Prophet scores of ≥0.35 (~10% error rate), ≥0.65 (~5% error rate), and ≥0.85 (~2% error rate). The data obtained after each of these filters are in Tables S1 and S2. For Tables S1 and S2, we used the data that was filtered using a Peptide Prophet score ≥0.35. Distributions for the case and control replicates were compared using an unpaired Student’s t test (with the replicate groups having equal sample size and unequal variance). Two tailed p values are reported.
Nondenaturing Coimmunoprecipitation
Cells were grown in 10 ml cultures to a density of ~1.8 × 107 cells/ml. Harvested cells were lysed in IP buffer. Lysates were diluted 1:5 in IP buffer and incubated for 16 hr at 4°C with 1:1,000 mouse anti-HSV antibody (Novagen) bound to 1.25 mg/ml Protein A Dynabeads (Invitrogen). Beads were washed three times in IP buffer. Proteins were eluted by incubation at 65°C for 10 min in SUMEB (SUME + 0.01% bromophenol blue). Proteins were separated on 8% SDS-PAGE gels, transferred to nitrocellulose, and visualized with anti-HSV (Novagen) or anti-HA (Sigma) antibodies.
Purification of Ubiquitinated Rpa190 from Yeast
Cells expressing 8His-ubiquitin and Rpa190-3HA were grown in 1 L cultures to a density of ~1.8 × 107 cells/ml. Harvested cells were lysed at 4°C in lysis buffer (20 mM Tris [pH 8.0], 200 mM NaCl with 1 mM PMSF and 10 mM NEM). The supernatant was removed and sonicated 4 × 20 s (duty cycle 60 and output 2). Lysates were clarified by centrifugation, and incubated with TALON resin (Novagen) for 4 hr at 4°C. The resin was washed 33 with wash buffer (20 mM Tris [pH 8.0], 200 mM NaCl, 1 mM PMSF, 10 mM NEM, 7.5 mM imidazole). Ubiquitinated proteins were eluted from the column by addition of 10 mM EDTA to the wash buffer. Anti-HA antibodies conjugated to Protein G Dynabeads (Dynal) were added to the ubiquitinated protein eluate and incubated overnight at 4°C. The beads were washed once in lysis buffer, and twice in IP wash buffer (100 mM Tris, 1 mM EDTA, 5% Glycerol, 1 mM DTT, 1 μM PMSF). The beads containing ubiquitinated Rpa190 were resuspended in IP wash buffer 2 and stored on ice.
Purification of Recombinant Ubp10 from Bacteria
A plasmid containing GST-TEV-UBP10 (gift from Ning Zheng, University of Washington) was transformed into T7 express cells (NEB). Cells were grown in lysogeny broth (LB) plus ampicillin to a density of ~1.0 × 107 cells/ml. Expression of GST-TEV-UBP10 was induced by addition of 250 mM IPTG (final concentration), followed by incubation overnight at 16°C. Harvested cells were lysed in lysis buffer (200 mM NaCl, 20 mM Tris [pH 8.0], 1 mM PMSF, 5 mM DTT). Clarified extract was applied to Glutathione Sepharose 4B resin (GE Healthcare). After washing, Ubp10 was cleaved from resin with recombinant TEV protease and concentrated using an Amicon Ultracel concentrator (cutoff 50 kDa).
In Vitro Rpa190 DUB Assay
Purified Ubp10 or vehicle was added to purified ubiquitinated Rpa190 and incubated for two hours at 37°C in reaction buffer (100 mM Tris, 1 mM EDTA, 5% Glycerol, 1 mM DTT, 1 μM PMSF). SUMEB was added to the samples, which were subsequently incubated at 65°C for 10 min. Proteins were separated on a 4%–12% Tris-Glycine gradient gel, transferred to nitrocellulose, and Rpa190-3HA visualized with anti-HA (Sigma) antibodies.
Polysome Analyses
Polysome analyses were performed similar to previously described (Steffen et al., 2008). Cultures grown to ~1 × 107 cells/ml were rapidly chilled with crushed, frozen YPD containing 100 μg/ml cycloheximide. All reagents were kept on ice and all steps performed at 4°C. Harvested cells were washed once with 10 ml lysis buffer (25 mM Tris-HCl [pH 7.5], 40 mM KCl, 7.5 mM MgCl2, 1 mM DTT, 0.5 mg/ml heparin, 100 μg/ml cycloheximide). Cells were resuspended in 1 ml lysis buffer and vortexed with glass beads to lyse. Detergents (1% final concentration of Triton X-100 and sodium deoxycholate) were added with vortexing and samples were incubated on ice for 5 min. Supernatants were clarified by centrifugation. Twenty A260 units of lysate in 1 ml total volume was loaded onto 11 ml linear 7%–47% sucrose gradients in 50 mM Tris-HCl (pH 7.5), 0.8 M KCl, 15 mM MgCl2, 0.5 mg/ml heparin, and 100 μg/ml cycloheximide. Gradients were centrifuged at 39,000 rpm at 4°C in a SW40 Ti swinging bucket rotor (Beckman) for 2 hr. Gradients were collected from the top and profiles read at 254 nm.
Northern Blot Analyses
Northern blot analyses were performed similarly to previously described (Pestov et al., 2008). Cultures were grown at 17°C in glucose medium for 72 hr for genetic depletion of the tagged protein. Cultures were grown, kept in log phase by frequent dilution with fresh media, to a final density of 0.53 – 0.87 × 107 cells/ml and harvested by centrifugation. Total RNA was extracted by the acid phenol method (Collart and Oliviero, 1994). RNA was resuspended in formamide loading dye and 3 μg was loaded per lane. Pre-RNAs were separated on an agarose-formaldehyde gel and transferred to a Hybond XL membrane. Methylene blue staining was used to detect the mature 18S and 25S rRNAs. Oligonucleotide probes used to detect pre-rRNAs are: b 5′GCT CTT TGC TCT TGC C, c 5′CCT CTG GGC CCC GAT TGC TCG AA, and e 5′GGC CAG CAA TTT CAA GT.
Cycloheximide-Chase Degradation Assays
Cycloheximide-chase degradation assays were performed similar to previously described (Gardner et al., 2005a). Cultures were grown to a cell density of 1 × 107 cells/ml. Cycloheximide was added to a final concentration of 50 μg/ml and the cells were further incubated at 25°C, 30°C, 33°C, or 38°C for 0–3 hr. In some cases, rapamycin was added (200 nM final concentration) or glucose or tryptophan was removed from the media 2 hr prior to cycloheximide addition. Cells were lysed at the appropriate time point in 200 μl SUMEB with 10 mM PMSF. Proteins were separated on 8% SDS-PAGE gels, transferred to nitrocellulose, and Rpa190-3HA visualized with anti-HA (Sigma) antibodies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Veronique Albanese, Judith Frydman, Vivian MacKay, and Kristan Steffen for assistance with polysome analyses, Joe Delaney for assistance with cell-cycle analyses, and Danesh Moazed for plasmids. This manuscript is in memory of Laura Sheard, whose generous help and thoughtful experimental insights helped us immeasurably with our studies. Support was from NIH Grants T32GM007750 (L.A.R. and B.J.R.), F31AA019842 (E.F.F.), T32HD07149 (E.F.F.), T32CA009259 (J.M.C.), R21RR025787 (R.G.G), R01GM052581 (S.J.B.), a Sidney Kimmel Scholar Award (R.G.G.), an Ellison Medical Foundation New Scholar Award in Aging (R.G.G.), and a Marian E. Smith Junior Faculty Award (R.G.G.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, two figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.07.009.
LICENSING INFORMATION This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
REFERENCES
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette JM, Baserga SJ. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010;16:2156–2169. doi: 10.1261/rna.2359810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Rockel T, Steinweger G, Hemmerich P, Risch J, von Mikecz A. Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol. Biol. Cell. 2002;13:3576–3587. doi: 10.1091/mbc.02-05-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system: from a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology. 2006;66(Suppl 1):S7–S19. doi: 10.1212/01.wnl.0000192261.02023.b8. [DOI] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. In: Preparation of yeast RNA. In Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. John Wiley & Sons; New York: 1994. pp. 13.12.11–13.12.12. [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Por-wancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Endo A, Kitamura N, Komada M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J. Biol. Chem. 2009a;284:27918–27923. doi: 10.1074/jbc.M109.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J. Cell Sci. 2009b;122:678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- Fátyol K, Grummt I. Proteasomal ATPases are associated with rDNA: the ubiquitin proteasome system plays a direct role in RNA polymerase I transcription. Biochim. Biophys. Acta. 2008;1779:850–859. doi: 10.1016/j.bbagrm.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005a;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 2005b;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grun-stein M, Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc. Natl. Acad. Sci. USA. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Kahana A, Gottschling DE. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6608–6620. doi: 10.1128/mcb.19.10.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts-Galbraith RH, Gould KL. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 2010;8:8. doi: 10.1371/journal.pbio.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol. Cell. 2008;30:498–506. doi: 10.1016/j.molcel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li J, Olson LM, Zhang Z, Li L, Bidder M, Nguyen L, Pfeifer J, Rader JS. Differential display identifies overexpression of the USP36 gene, encoding a deubiquitinating enzyme, in ovarian cancer. Int. J. Med. Sci. 2008;5:133–142. doi: 10.7150/ijms.5.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 2009;16:740–746. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YH, Charette JM, Baserga SJ. Assembling a protein-protein interaction map of the SSU processome from existing datasets. PLoS ONE. 2011;6:e17701. doi: 10.1371/journal.pone.0017701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S, Gouy M, Marck C, Sentenac A, Buhler JM. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J. Biol. Chem. 1988;263:2830–2839. [PubMed] [Google Scholar]
- Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol. 2008;22:22–11. doi: 10.1002/0471143030.cb2211s39. [DOI] [PubMed] [Google Scholar]
- Philippi A, Steinbauer R, Reiter A, Fath S, Leger-Silvestre I, Milkereit P, Griesenbeck J, Tschochner H. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 2010;38:5315–5326. doi: 10.1093/nar/gkq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps KR, Charette JM, Baserga SJ. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip Rev RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Rauch A, Bellew M, Eng J, Fitzgibbon M, Holzman T, Hussey P, Igra M, Maclean B, Lin CW, Detter A, et al. Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J. Proteome Res. 2006;5:112–121. doi: 10.1021/pr0503533. [DOI] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, Gardner RG. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493:176–184. doi: 10.1016/j.gene.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multi-ubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Kawasaki M, Dundr M, Koberna K, Müller WG, Tsujimura-Takahashi T, Komatsu W, Hayano T, Isobe T, Raska I, et al. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol. Cell. Biol. 2006;26:5131–5145. doi: 10.1128/MCB.02227-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BW, Toews J, Kast J. Utility of formaldehyde crosslinking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- Yamamoto RT, Nogi Y, Dodd JA, Nomura M. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.