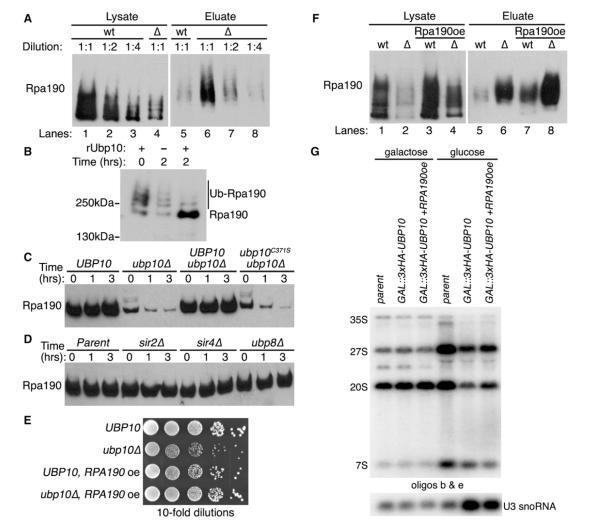
Figure 4. Ubp10 Controls Rpa190 Ubiquitination and Stability.
(A) Ubiquitin proteomes from UBP10 (wt) or ubp10Δ (Δ) cells were isolated by metal affinity purification. Levels of Rpa190-3HA in lysates (total protein) and eluates (ubiquitinated Rpa190) were determined by western analysis using anti-HA antibodies. Lanes 1 and 4 indicate steady-state levels of Rpa190 in total lysates of UBP10 and ubp10Δ cells. Lanes 5 and 6 show levels of ubiquitinated Rpa190 in the ubiquitin proteome of UBP10 and ubp10Δ cells.
(B) In vitro Rpa190 deubiquitination assay. Recombinant Ubp10 was added to purified ubiquitinated Rpa190 for the indicated time. Western analysis of Rpa190 was performed using anti-HA antibodies.
(C) Cycloheximide-chase degradation assays of UBP10, ubp10Δ, UBP10-3HSV ubp10Δ, or ubp10C371S-3HSV ubp10Δ cells expressing Rpa190-3HA. Time after cycloheximide addition is indicated above each lane. Western analysis of whole-cell extracts was performed using anti-HA antibodies.
(D) Cycloheximide-chase degradation assays of UBP10, sir2Δ, sir4Δ, or ubp8Δ cells expressing Rpa190-3HA. Assay performed as in (C).
(E) Growth of UBP10 and ubp10Δ cells with or without RPA190 overexpression (oe). Tenfold serial dilutions of cells were spotted onto the appropriate media and incubated at 30°C for 3 days.
(F) Ubiquitin proteome analysis from UBP10 (wt) or ubp10Δ (Δ) cells overexpressing (oe) Rpa190 was performed as in (A).
(G) RPA190 overexpression partially rescues the decrease in pre-rRNA species when UBP10 is genetically depleted. Total RNA was extracted from yeast in which the indicated protein was genetically depleted for 72 hr at 17°C. Pre-rRNA species were detected by northern analysis using the indicated oligonucleotide probes from Figure 2B.