FIGURE 6:
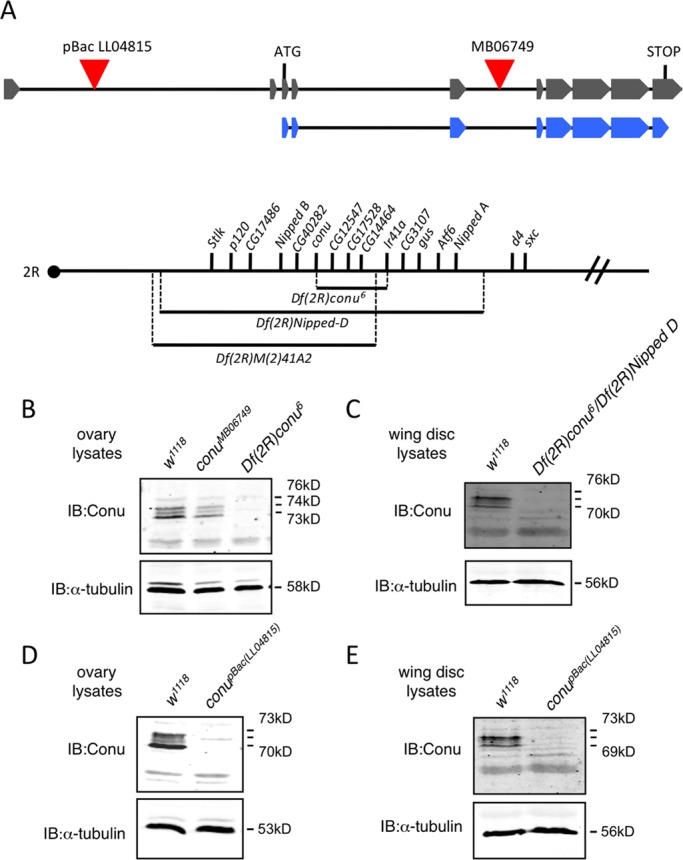
Conu is not required for viability. (A) A schematic of the predicted conu gene transcript RA is shown in gray, with boxes representing exons and lines representing introns. The schematic is not to scale, but relative distances are shown. The coding region is shown underneath in blue. A piggyBac element (LL04815) is inserted in an intron 5′ to the start of translation, while a Minos element (MB06749) is inserted in an intron between coding exons 3 and 4, as indicated by the red triangles. The two deficiencies used to screen the Minos element excision lines (Df(2R)Nipped-D and Df(2R)M(2)41A2) are indicated, along with the small deletion (Df(2R)conu6) produced by excision of the Minos element, with the genes uncovered by each indicated above as reported by Myster et al. (2004) and this study. The dot represents the centromere, and genes are shown in order on the chromosome, but distances between genes are not drawn to scale. (B) Ovary lysates from w1118, conuMB06749, and Df(2R)conu6 homozygous adult females showed that while Conu is expressed at similar levels in conuMB06749 and w1118 ovaries, little or no protein is present in Df(2R)conu6 ovary lysates. (C) Wing disk protein lysates from w1118 and transheterozygous Df(2R)conu6/Df(2R)Nipped-D animals revealed little or no Conu present in transheterozygous animals. (D and E) Little or no Conu is present in ovary or wing disk lysates from animals with a piggyBac insertion (LL04815), inserted 5′ to the conu translation start site. α-tubulin was used as a loading control for all samples analyzed.
