Figure 2. Base-pairing formation of the predicted hairpin stem is required for efficient −1 PRF attenuation activity.
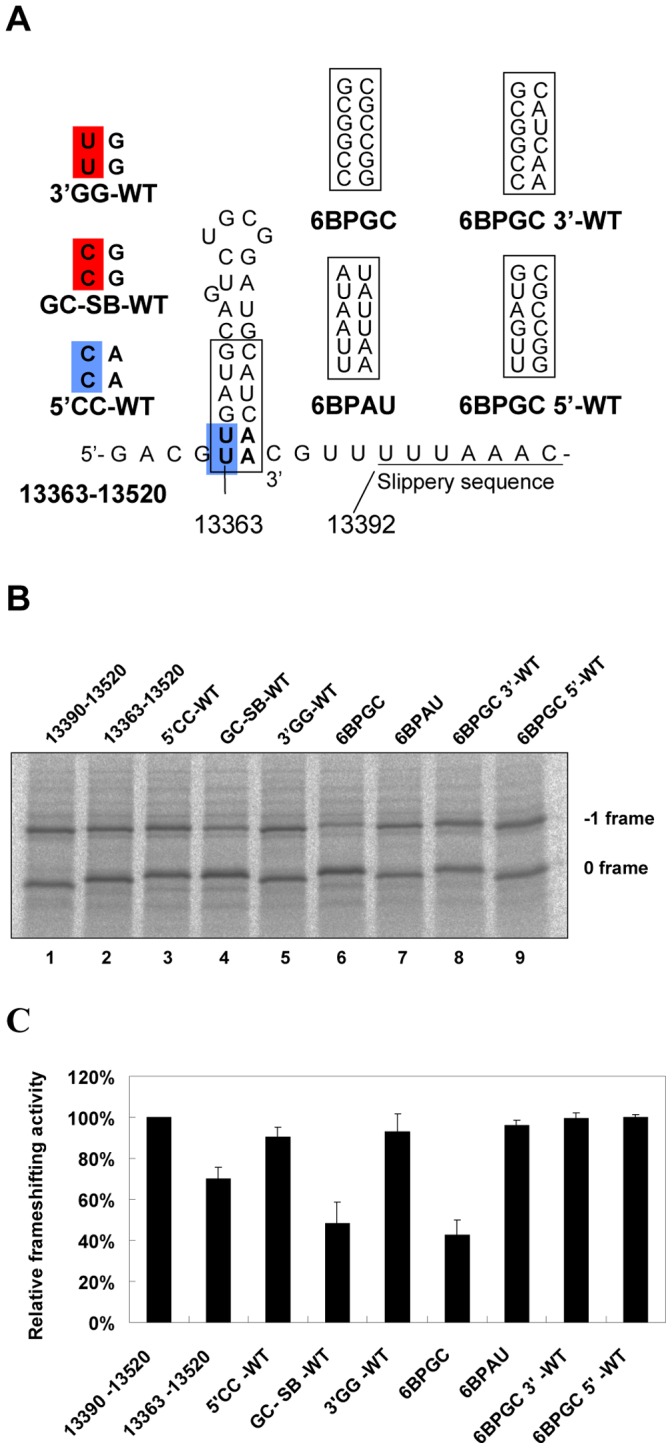
(A) Illustration of constructs with different base-pairing schemes at the lower stem of the predicted hairpin. For each mutant, the nucleotide composition after mutation is boxed or boldly typed. Two nucleotides, 27 nucleotides upstream of the 0-frame E site and those involved in terminal stem base-pairing formation, are colored for comparison. (B) In vitro −1 PRF assays by SDS-PAGE analysis of 35S methionine-labeled translation products for constructs with different base-pairing schemes. The 0 and −1 frame products are labeled as indicated. (C) Relative frameshifting activity of (B) with the frameshifting efficiency of construct 13390–13520 as 100% (for comparison purposes). Value for each construct was the mean of three independent experiments, with the bar representing the standard error of the mean.
