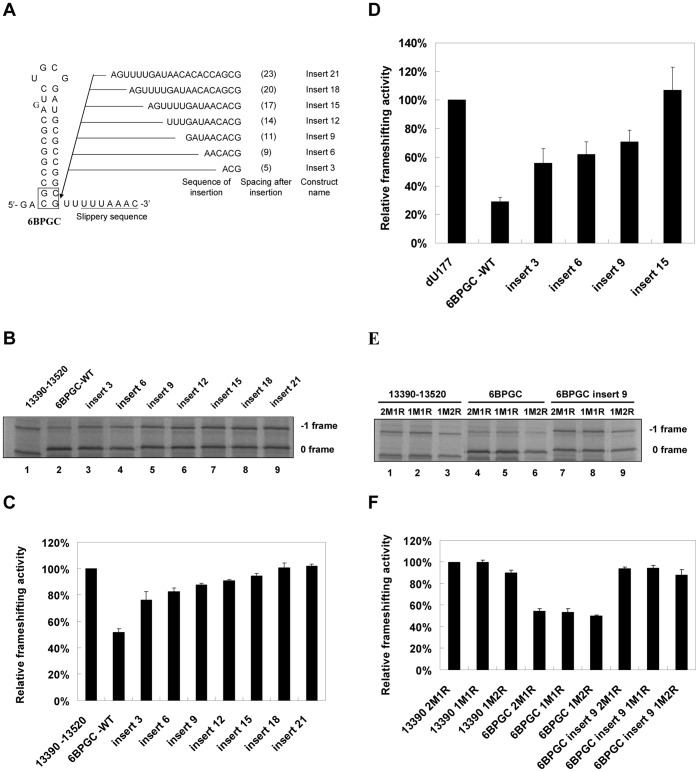
Figure 5. Proximity to the slippery site determines the attenuation potency of a hairpin.
(A) Scheme showing the inserted nucleotide sequences between the extended base of the 6BPGC hairpin stem and the slippery site with spacing numbers after insertion shown in parentheses. The two extended base pairs involving spacer are boxed. The inserted sequences were designed to retain no stable secondary structure and regenerate the 3′-flanking CGUU sequence to prevent flanking sequence effects affecting frameshifting efficiency [12], [13]. (B) In vitro −1 PRF assays by SDS-PAGE analysis of 35S methionine-labeled translation products for constructs with insertions as shown in (A). (C) Relative frameshifting activity with that of the construct 13390–13520 being treated as 100%. Error bars, s.d.; n = 3. (D) Relative −1 frameshifting activity based on dual-luciferase assays from yeast cells, transfected with reporters containing selected insertion mutants in (A) and SARS-PK replaced by DU177 pseudoknot. Frameshifting efficiency of the construct containing the DU177 pseudoknot alone was treated as 100% (for comparison purposes). Error bars, s.d.; n = 3. (E) In vitro −1 PRF assays by SDS-PAGE analysis for reporter constructs (with or without nucleotide insertion in the region between the extended base of 6BPGC hairpin stem and the slippery site) under different conditions with variations in relative amounts of mRNA and Retic lysate. Condition designations: 2M1R (mRNA 100 ng/Retic lysate 1.7 μl); 1M1R (mRNA 50 ng/Retic lysate 1.7 μl); 1M2R (mRNA 50 ng/Retic lysate 3.4 μl) in a total of 5 μl/reaction. (F) Relative frameshifting activity of (E) using frameshifting efficiency of 13390–13520 construct in 2M1R condition as 100%. Error bars, s.d.; n = 3.