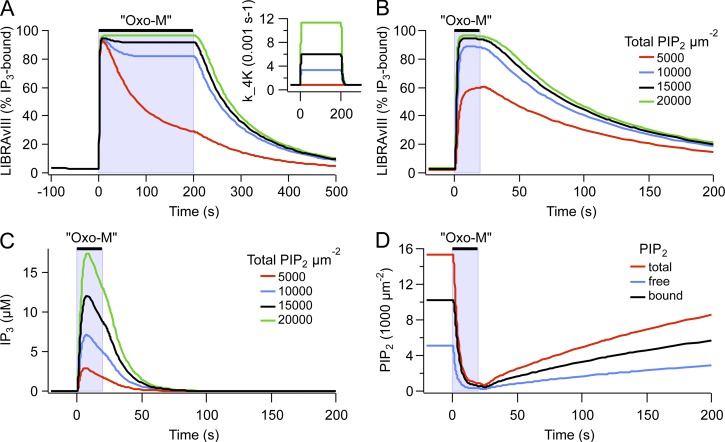
Figure 6.
Modeling results suggesting a pool of reversibly bound PIP2. (A) Model predictions of the response of LIBRAvIII (percentage of IP3 bound) after a 200-s application of 10 µM Oxo-M with or without 4-kinase acceleration. Inset shows the different rates of 4-kinase activity during Oxo-M. (B) Model simulation showing the response of LIBRAvIII (percentage of IP3 bound) after a 20-s application of 10 µM Oxo-M computed for different values of the parameter fold_PIP2 (1, 2, 3, 4) and therefore different amounts of bound PIP2 (0, 5,000, 10,000, and 15,000 µm−2). Free PIP2 was kept at 5,000 µm−2. (C) Model prediction of the concentration of IP3 produced after a 20-s application of Oxo-M computed for different values of the parameter fold_PIP2 as in C. (D) Model prediction of the response of free, bound, and total (free plus bound) PIP2 after a 20-s exposure to 10 µM Oxo-M with fold_PIP2 = 3. [LIBRAvIII] was 6 µM for A and B and 0 µM for C and D. To maintain the kinetic properties of the model, the rate constants for all reactions of PI metabolism (4-K, 4-P, 5-K, 5-P, and PLC) were scaled by fold_PIP2 (see Tables 1 and 2).