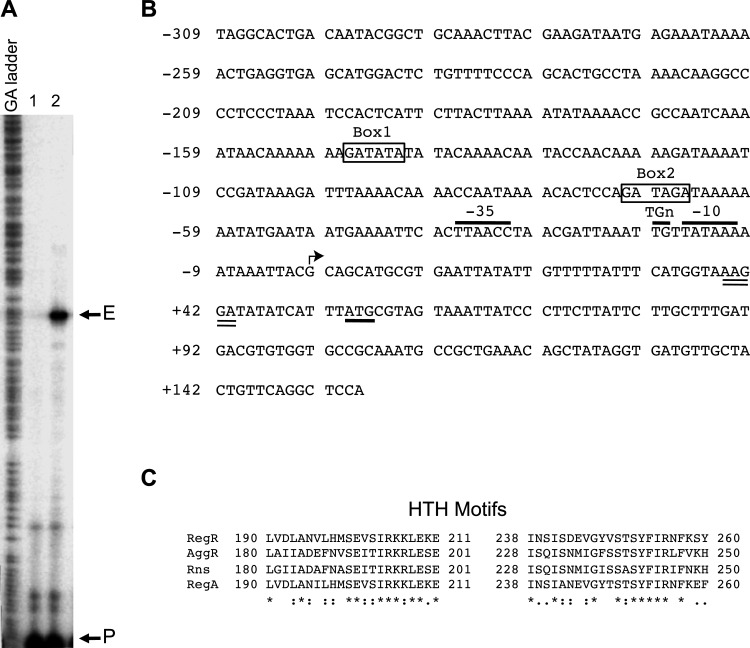
Fig 1.
The sefA regulatory region and the double helix-turn-helix DNA-binding motifs of RegR and its homologs. (A) The start site of transcription of the sefA promoter was mapped by primer extension using RNA isolated from E. coli MC4100 strains containing pACYC184-regR (pYS3) with either pMU2385 (control) or sefA-lacZ1 (pYS9). Lane 1, control experiment using RNA from E. coli MC4100 containing pACYC184-regR and pMU2385. Lane 2, experiment using RNA from E. coli MC4100 containing pACYC184-regR and sefA-lacZ1. The positions corresponding to the primer and the extension product are marked with P and E, respectively. (B) Nucleotide sequence of the sefA regulatory region. The numbering on the left of the sequence is relative to the transcriptional start site of sefA, which is indicated by an angled arrow. The putative −10 region, TGn motif, and −35 region are indicated and overlined. The putative start codon and ribosome-binding site are underlined and double underlined, respectively. The putative RegR-binding sites, Box1 and Box2, are boxed and labeled. (C) Alignment of helix-turn-helix (HTH) motifs of RegR, AggR, Rns, and RegA was done using the PS01124 profile defined in PROSITE (65). The numbers flanking the sequences indicate the positions of amino acid residues of the regulatory proteins. Identical and conserved amino acids are indicated with asterisks and colons, respectively.