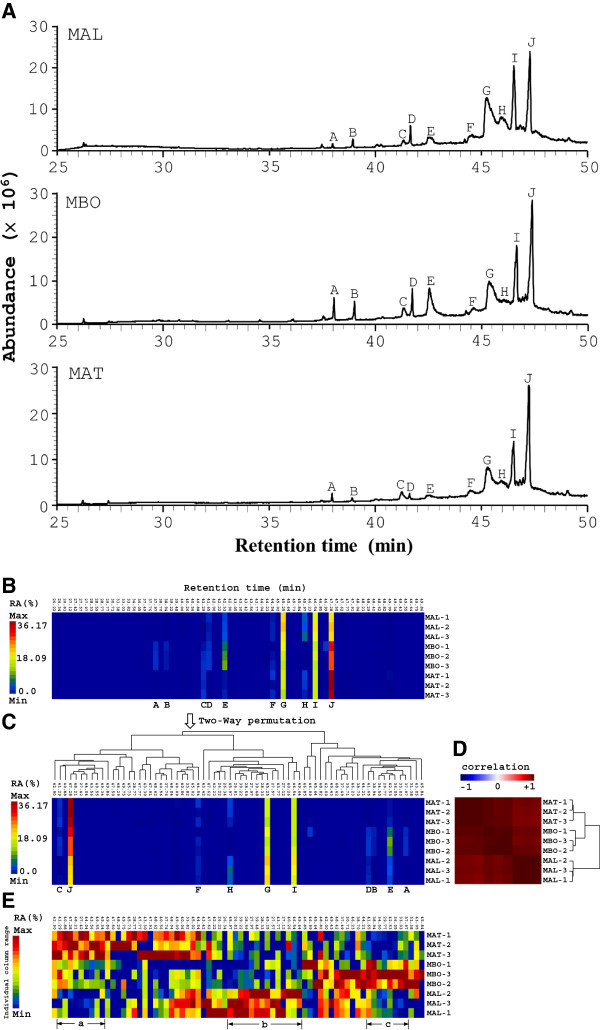
Figure 1.
GC-MS profiles and cluster analysis of the three Morus species. (A) Branches of M. alba (MAL), M. atropurpurea (MAT), and M. bombycis (MBO) underwent supercritical CO2 extraction and GC-MS analysis at a dose of 10 mg/ml. One representative GC-MS chromatogram from 3 batches of each plant extract is shown. The sizes of the various GC-MS peaks are measured as percent relative area (RA (%)) of the largest GC-MS peak in the chromatogram. (B) Matrix visualization with hierarchical cluster trees (HCT) for GC-MS profiles of the Morus species. A representative GC-MS profile of the 3 Morus species from Figure 1 is displayed as a matrix map with 9 rows, each representing one sample, and 70 columns, each representing one retention time. A rainbow spectrum was used to color code the size of the chromatographic peaks (expressed in percent relative area, RA (%)) in the whole matrix. Red denotes the largest peak (maximum RA (%)) and blue denotes the smallest peak (minimum RA (%)). (C) Two-way permuted data matrix of (B). The tree structure above the data matrix represents average linkage hierarchical cluster tree (HCT) for 70 different retention times. (D) Map of correlation matrix for the 9 samples (three samples per species). A bi-directional blue–white–red spectrum is used to denote negative-to-positive correlation. The tree structure besides the correlation matrix represents average linkage HCT for 9 samples. (E) Identical data matrix map as in (B) except each column is colored using a rainbow color spectrum scaled individually for that particular column instead of a rainbow color spectrum scaled for all columns in the matrix.