Abstract
Background
Pepino mosaic, once an emerging disease a decade ago, has become endemic on greenhouse tomatoes worldwide in recent years. Three distinct genotypes of Pepino mosaic virus (PepMV), including EU, US1 and CH2 have been recognized. Our earlier study conducted in 2006–2007 demonstrated a predominant EU genotype in Canada and United States. The objective of the present study was to monitor the dynamic of PepMV genetic composition and its current status in North America.
Results
Through yearly monitoring efforts in 2009–2012, we detected a dramatic shift in the prevalent genotype of PepMV from the genotype EU to CH2 in North America since early 2010, with another shift from CH2 to US1 occurring in Mexico only two years later. Through genetic diversity analysis using the coat protein gene, such genotype shifting of PepMV in North America was linked to the positive identification of similar sequence variants in two different commercial tomato seed sources used for scion and rootstock, respectively. To allow for a quick identification, a reverse transcription loop-mediated isothermal amplification (RT-LAMP) system was developed and demonstrated to achieve a rapid identification for each of the three genotypes of PepMV, EU, US1 and CH2.
Conclusion
Through systemic yearly monitoring and genetic diversity analysis, we identified a linkage between the field epidemic isolates and those from commercial tomato seed lots as the likely sources of initial PepMV inoculum that resulted in genetic shifting as observed on greenhouse tomatoes in North America. Application of the genotype-specific RT-LAMP system would allow growers to efficiently determine the genetic diversity on their crops.
Keywords: Pepino mosaic virus, Genetic diversity, Genotype shift, Loop-mediated isothermal amplification
Background
Tomato (Solanum lycopersicum L.), an economically important vegetable, is widely grown around the world with leading tomato producing countries in China, the United States, Italy and Spain [1]. Although most of tomatoes are field-grown, protected production systems (greenhouse) have increased significantly in recent years, including in North America [2]. Pepino mosaic is one of several emerging viral diseases that seriously constrain the profitable production of tomatoes worldwide [3].
Pepino mosaic virus (PepMV) from the genus Potexvirus in the family Alphaflexiviridae was first reported to infect pepino (S. muricatum L.) in 1980 [4]. This virus was not considered an issue until 20 years later when it was first reported to infect greenhouse tomatoes in the Netherlands [5]. Since then, the disease has become widespread in Europe [6-14]. PepMV has also caused serious problems on greenhouse tomatoes in North America [15-18] and in South America [19-21]. Furthermore, this virus was detected in China [22], Syria [23], and South Africa [24]. Thus, PepMV has become one of the most economically important viruses infecting greenhouse tomatoes worldwide [3,25,26]. The typical disease symptoms on tomatoes include mosaic, yellow patches, necrotic lesions and uneven ripening of fruits resulting in marbling or flaming in appearance on mature fruits. It was estimated that the poor quality of PepMV-infected fruits could reduce tomato market value by up to 36% [27].
Three major genotypes of PepMV (namely EU, US1 and CH2), which share only 78-82% genomic nucleotide sequence identity, have been reported [9,10,16,21,25,26]. Even though a greater level of PepMV genetic diversity was initially recognized in the American continents [16,21], further population genetic analysis in Canada and the U.S. showed a predominant EU genotype, with only a small fraction of field isolates in US1 and CH2 genotypes [17]. However, several studies in Europe in recent years demonstrated that the prevalent PepMV genotype has shifted from a predominant EU [11,28] to CH2 [14,29-31]. Recent outbreaks of pepino disease in association with the CH2 genotype in broader geographic regions in the world, including the Middle East [23] and South Africa [24], raised concerns of possible seed transmission. Although PepMV on tomato is localized on seed coat (testa) and not in embryo, mechanical transmission from a contaminated seed could easily induce a new infection [32]. A low rate (0.026%) of PepMV seed transmission in tomato has already been observed [33]. These results on seed transmission demonstrated the importance of selecting and planting tomatoes from PepMV-tested negative seed lots. With intensive cultural practices, as required under a greenhouse tomato production system (i.e., grafting, de-leafing, inter-cropping and bumble bee pollination), even with a small number of PepMV-infected plants serving as sources of initial inocula could result in a serious disease epidemic. Thus, a timely detection and rapid genotype determination would be a prerequisite for deploying effective strategies in disease management.
Currently, several molecular-based methods have been developed to determine the genetic diversity of PepMV, including restriction fragment length polymorphism (RFLP) [34,35], reverse transcription- polymerase chain reaction (RT-PCR) and sequencing [14,17], real-time RT-PCR [36], and hybridization [30]. All such molecular methods required some specialized instruments or sophisticated laboratory conditions, making it difficult to apply for field analysis. With recent trends in genotype shifting in Europe, growers in North America are concerned about their crop’s health status and would request additional sequencing to determine the specific genotype [17]. Although this method is accurate, it is costly and time consuming. Thus, we were interested in developing a rather simple and quick reverse transcription loop-mediated isothermal amplification (RT-LAMP) for efficient identification of PepMV genotypes. In recent years, the RT-LAMP technology [37,38] has been successfully applied for plant virus and viroid detection [39-47]. In an effort to understand the current genetic diversity and evolution of PepMV in North America, we established a surveillance system with assistance from participating growers in Canada, Mexico and the U.S.A. on sample collection. We developed and used LAMP to monitor the presence and distribution of PepMV genetic diversity in North America.
Results and discussion
Development and validation of genotype-specific RT-PCR for PepMV
Through direct sequencing of RT-PCR amplified products and sequence analysis, specificity of the newly designed primers (Table 1) for genotype-specific amplification was evaluated on 50 field samples collected in North America in 2012 (Table 2). All 24 samples collected in Texas, USA were positive for CH2, while 10 of these samples were also of mixed infection with EU genotype but negative for US1. The 16 samples from Jocotitlan, Mexico were infected by the US1 genotype. Two among them were also of mixed infection with CH2 genotype. All 10 samples from British Columbia, Canada, were positive for CH2 by RT-PCR (Table 2).
Table 1.
Primer sequences for Pepino mosaic virus genotype-specific loop mediated isothermal amplification and RT-PCR
| Genotype | Primer designation | Sequence* | Position on genome | GenBank accession # |
|---|---|---|---|---|
| Primers for genotype-specific RT-LAMP | ||||
| CH2 |
CH2_F3 |
5′-CGATGAAGCTGAACAACATTTCC-3′ |
4297-4319 |
DQ000985 |
| CH2_FIP |
5′-CTTAATGGGTTGATCTTGGTGGAAGCTGTGAGAAAGCTTCACAAAC-3′ |
4403-4381 + 4321-4343 |
||
| CH2_BIP |
5′-GGGTTAAGTTTTCCCCAGTTTGAAAATTCCTTCAGTGTTAATCTTGTG-3′ |
4404-4428 + 4498-4476 |
||
| CH2_B3 |
5′-TCCAGCAATTCCGTGCACAACAA-3′ |
4523-4501 |
||
| CH2_Loop F |
5′-GGCCTCGCCTTGATGGA-3′ |
4360-4344 |
||
| CH2_Loop B |
5′-TGGAAAGATCAACTTTGATCAATT-3′ |
4429-4452 |
||
| EU |
EU_F3 |
5′-ACCAAGAAGATACAAAATTTGC-3′ |
6090-6111 |
FJ940223 |
| EU_FIP |
5′-TRAGACCATCAGCAGGCTGC TGCATTTGACTTCTTCGATG-3′ |
6173-6154 + 6112-6131 |
||
| EU_BIP |
5′-TCAGGCARCCAAATGAGAAAGAAACCTGTGGAGATCTTTTGC-3′ |
6174-6196 + 6256-6238 |
||
| EU_B3 |
5′-TGACTTCTCCAAGTGTGG-3′ |
6284-6267 |
||
| EU_Loop F |
5′-TGGCAGGGTTGGTGACTC-3′ |
6149-6132 |
||
| EU_Loop B |
5′-CTAGCTGCTCACTCCGTAGCTAA-3′ |
6197-6219 |
||
| US1 |
US1_F3 |
5′-GCATTCATACCAAATGGGAG-3′ |
4255-4274 |
FJ940225 |
| US1_ FIP |
5′-TGCGAACAGCCAAGAAATGT-ATAAATTGCATGAATACCTTACTCC-3′ |
4334-4315 + 4275-4299 |
||
| US1_ BIP |
5′-TTGCACAAACTCCACCAAGGACTTAACCCGTCAATGTGTT-3′ |
4337-4356 + 4415-4396 |
||
| US1_ B3 |
5′-CCATTTCGAACAGGGGAA-3′ |
4433-4416 |
||
| US1_ Loop F |
5′-TGCTCAGCTTCATCA-3′ |
4413-4299 |
||
| US1_ Loop B |
5′-TGAAGCCATGAGACTT-3′ |
4357-4372 |
||
| Primers for genotype-specific RT-PCR | ||||
| CH2 |
PeppMVCH2CPF |
5′- caggaaacagctatgacGTTTTCCTCAATTGTGAAAT-3′ |
5568-5587 |
DQ000985 |
| PepMVCH2CPR |
5′- tgtaaaacgacggccagtTTTTTTTTTTATTTAGTAGATTTAGATAC-3′ |
6412-6394 |
||
| EU |
PepMVEUCPF |
5′ – caggaaacagctatgacGTTTTCCTAAATTTGAAAAT-3′ |
5572-5591 |
FJ940223 |
| PepMVEUCPR |
5′ – tgtaaaacgacggccagtATTTCAAAGAAATAATTAGG-3′ |
6410-6391 |
||
| US1 |
PepMVUS1CPF |
5′ - caggaaacagctatgacGTTTTCCTAGTGTTTGAAA-3′ |
5570-5588 |
FJ940225 |
| PepMVUS1CPR |
5′ - tgtaaaacgacggccagtAAATTACAAAAGCAATTTATTG-3′ |
6415-6394 |
||
| Primers for full genome sequencing | ||||
| CH2 | KL11-124:CH2-1F |
5′- caggaaacagctatgacGAAAACAAAACATAACACATAATATC-3′ |
1-26 |
DQ000985 |
| KL11-131: CH2-1226R |
5′- tgtaaaacgacggccagtTCATGCACCTCCAGTCATGT-3′ |
1226-1207 |
||
| KL11-125: CH2-902F |
5′- caggaaacagctatgacAAAAATAGCTTTGTGACCTTTCC-3′ |
902-924 |
||
| KL11-132: CH2-1741R |
5′- tgtaaaacgacggccagtGCTGGAAGTGTCAGATGCAA-3′ |
1741-1722 |
||
| KL11-126: CH2-1395F |
5′- caggaaacagctatgacCCAATTTAGTCAAACAAGGCGTA-3′ |
1395-1417 |
||
| KL11-133: CH2- 2719R |
5′- tgtaaaacgacggccagtAATTGGCACTTTGCACTTTTG-3′ |
2719-2699 |
||
| KL11-127: CH2-2383F |
5′- caggaaacagctatgacGATTCAACCTGGCTTTCCAA-3′ |
2383-2402 |
||
| KL11-134: CH2- 3738R |
5′- tgtaaaacgacggccagtGTTTGGGCGGTTCTGTTAAA-3′ |
3738-3719 |
||
| KL11-128: CH2- 3379F |
5′- caggaaacagctatgaCGACCTGGGAGATTTGTGCTG-3′ |
3379-3398 |
||
| KL11-135: CH2- 4734R |
5′- tgtaaaacgacggccagtGACAGGGGTCACCAAAAATG-3 |
4734-4715 |
||
| KL11-129: CH2- 4381F |
5′- caggaaacagctatgacTCCACCAAGATCAACCCATT-3′ |
4381-4400 |
||
| KL11-136: CH2- 5748R |
5′- tgtaaaacgacggccagtAAATCACTTAGGGAAGGAGCTG-3′ |
5748-5727 |
||
| KL11-130: CH2- 5405F |
5′- caggaaacagctatgacGCCGTAATATTCACCAGCATC-3′ |
5405-5425 |
||
| KL11-137: CH2-6412 | 5′- tgtaaaacgacggccagtTTTTTTTTTTATTTAGTAGATTTAGATAC-3′ | 6412-6394 | ||
*Primer adapters (M13 forward and reverse primer sequences, in lowercase) were added to specific viral sequences to facilitate direct sequencing of the amplified RT-PCR products.
Table 2.
Genotyping of Pepino mosaic virus isolates with reverse transcription loop-mediated isothermal amplification (RT-LAMP) and its validation with genotype-specific RT-PCR
| Country of origin | Isolate name | ELISAa |
EU |
US1 |
CH2 |
|---|---|---|---|---|---|
| RT-PCR/RT-LAMP | RT-PCR/RT-LAMP | RT-PCR/RT-LAMP | |||
| USA |
VFTX12-01 |
(++++) |
(−)/(+)b |
−/− |
+/+ |
| VFTX12-02 |
(++++) |
(−)/(+)b |
−/− |
+/+ |
|
| VFTX12-03 |
(++++) |
(−)/(+)b |
−/− |
+/+ |
|
| VFTX12-04 |
(++++) |
−/− |
−/− |
+/+ |
|
| VFTX12-05 |
(++++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-06 |
(++++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-07 |
(++++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-08 |
(++++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-09 |
(+++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-10 |
(++++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-11 |
(++++) |
−/− |
−/− |
+/+ |
|
| VFTX12-12 |
(++++) |
−/− |
−/− |
+/+ |
|
| VFTX12-13 |
(+++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-14 |
(+++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-15 |
(+++) |
+/+ |
−/− |
+/+ |
|
| VFTX12-16 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-18 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-19 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-20 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-21 |
(+) |
+/+ |
−/− |
+/+ |
|
| VFTX12-22 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-23 |
(+) |
−/− |
−/− |
+/+ |
|
| VFTX12-24 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFTX12-25 |
(+++) |
−/− |
−/− |
+/+ |
|
| Mexico |
BNMX12-01 |
(+++) |
−/− |
+/+ |
−/− |
| BNMX12-02 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-03 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-04 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-05 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-06 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-07 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-08 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-09 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-10 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-11 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-12 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-13 |
(+++) |
−/− |
+/+ |
+/+ |
|
| BNMX12-14 |
(+++) |
−/− |
+/+ |
−/− |
|
| BNMX12-15 |
(++) |
−/− |
+/+ |
−/− |
|
| BNMX12-16 |
(++) |
−/− |
+/+ |
+/+ |
|
| Canada |
VFBC12-01 |
(+++) |
−/− |
(−)/(+)b |
+/+ |
| VFBC12-02 |
(++++) |
−/− |
(−)/(+)b |
+/+ |
|
| VFBC12-03 |
(+++) |
−/− |
(−)/(+)b |
+/+ |
|
| VFBC12-04 |
(−) |
−/− |
−/− |
+/+ |
|
| VFBC12-05 |
(+++) |
−/− |
−/− |
+/+ |
|
| VFBC12-06 |
(++) |
−/− |
−/− |
+/+ |
|
| VFBC12-07 |
(+) |
−/− |
−/− |
+/+ |
|
| VFBC12-08 |
(++++) |
−/− |
−/− |
+/+ |
|
| VFBC12-09 |
(+) |
−/− |
−/− |
+/+ |
|
| VFBC12-10 |
(+++) |
−/− |
−/− |
(+)/+c |
|
| Positive |
|
(+++) |
+/+ |
+/+ |
+/+ |
| Negative | (−) | −/− | −/− | −/− |
a ELISA ratings: (−) <0.100; (+): 0.101-0.500; (++): 0.501-1.000; (+++): 1.001-2.000; (++++) >2.001.
b A discrepancy was observed in these isolates with a negative (−) RT-PCR, but a positive (+) RT-LAMP. A confirmation test through sequencing of cloned RT-LAMP products was performed to determine the authenticity of their viral origin.
c The result for a positive (+) RT-PCR was not consistent from one experiment to another while RT-LAMP was consistently positive.
The sensitivity and reliability of RT-LAMP in genotype determination of PepMV isolates
The specificity of the developed RT-LAMP for each genotype (including EU, US1 and CH2) was validated with a genotype-specific RT-PCR (Figure 1 and Table 2). A greater sensitivity was obtained for RT-LAMP than that of its respective RT-PCR, as demonstrated with two CH2 isolates (Figure 1). Consequently, these genotype-specific RT-LAMPs were used to evaluate field samples for their genotype composition, confirming a single or a mixed infection of two or three genotypes (Table 2). Although test results using genotype-specific RT-LAMP and RT-PCR on 44 of 50 field collected samples were in full agreement with one another, there were some minor discrepancies in six other samples (Table 2). RT-LAMP was able to detect the presence of genotypes CH2 and EU sequences in three Texas samples (isolates, VFTX12-01 to VFTX12-03); whilst only CH2 was detectable by RT-PCR in the same samples. Similarly, both CH2 and US1 sequences could be identified by RT-LAMP in three Canadian samples (isolates: VFBC12-01 to VFBC12-03); RT-PCR, however, only detected the presence of genotype CH2. To resolve such discrepancies, sequences from cloned RT-LAMP products were obtained and validated. Thus, RT-LAMP demonstrated a greater sensitivity than RT-PCR in detecting such low concentration of the virus.
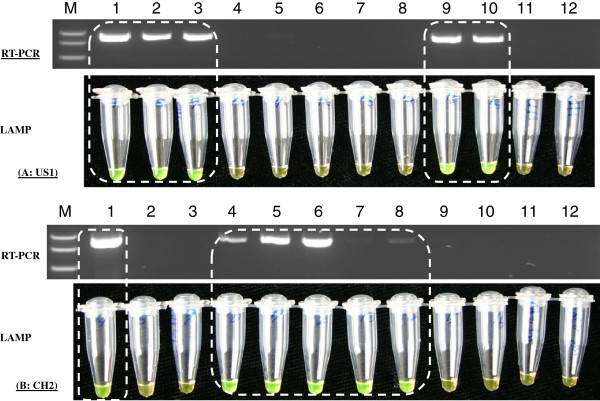
Figure 1.
Comparative analysis of a genotype-specific detection of Pepino mosaic virus isolates using RT-LAMP and RT-PCR. The upper panel (A) was for genotype US1 isolates. The lower panel (B) was for genotype CH2 isolates. Lane M: PCR marker; Lane 1 was a positive control in a mixed infection with two genotypes of PepMV (US1 and CH2); Lanes 2–10 were 9 field samples infected with PepMV; Lane 11 was a health tomato (negative) control; Lane 12 was a non-template (blank) control. PepMV isolates in Lanes 2, 3, 9 and 10 were typed as US1. The isolates in Lanes 4–8 were shown to be CH2 type. Note, the faint products in lanes 7–8 by RT-PCR, were clearly visible by RT-LAMP.
The prevalent PepMV genotype shifted in North America
PepMV isolates collected from selected greenhouses in Canada, Mexico and the U.S. were genotyped over seven consecutive years (2006–2012). Prior reported genotyping data from an imported seed sample from Chile [21] and the survey and genotyping results in 2006–2007 [17] served as base lines to evaluate the dynamic of genotype shift in North America. Prior to 2009, prevalent isolates of PepMV in Canada and the U.S. were predominantly in the EU genotype. Surprisingly, a shift in the prevalent genotype was observed in early 2010 when all 10 samples (four from Canada, three from Mexico and three from the U.S.) were shown to be CH2. Six additional samples (five from Mexico and one from the U.S.) collected in 2011 also belonged to genotype CH2 (Table 3). To better assess the genetic diversity of PepMV, a larger number of samples were collected and analyzed in 2012; 10, 16, and 32 samples were collected from Canada, Mexico, and the U.S. greenhouses, respectively. Greater variations in the genetic diversity were observed in the three countries (Table 3). CH2 sequences were detected in all 10 samples from Canada. Among them, two were in a mixed infection of EU. In the U.S., 31 of 32 samples analyzed contained the CH2 genotype sequence, with 17 in a single infection and 14 a mixed infection of CH2 and EU. Only 1 of 32 samples was singly infected by EU (Table 3). In contrast, 16 of 22 samples collected from Mexico in 2012 were genotyped as US1, with 14 of them containing a single infection and the other two a mixed infection of US1 and CH2 (Table 2). These results indicated a genotype shift from CH2 to US1 in Mexico. All sequence data were deposited in GenBank [JX866611 - JX866663].
Table 3.
Genetic diversity and dynamic of Pepino mosaic virus genotypes in North America from 2006 to 2012
| Year | Canadaa | Mexico | U.S.A. | Reference |
|---|---|---|---|---|
| 2006 |
EU (4/4) |
ntb |
EU (7/7) |
Ling et al., 2008 [17] |
| 2007 |
nt |
nt |
EU + CH2 (1/1) |
|
| 2009 |
nt |
0/10 |
EU (4/4) |
This study |
| 2010 |
CH2 (4/4) |
CH2 (3/3) |
CH2 (3/3) |
|
| 2011 |
nt |
CH2 (5/5) |
CH2 (1/1) |
|
| 2012 | CH2 (7/10), US1 + CH2 (3/10) | US1 (14/22), CH2 (6/22), US1 + CH2 (2/22) | CH2 (17/32), CH2 + EU (14/32), EU (1/32) |
a Greenhouse location: Canada (Delta, British Columbia), Mexico (Jocotitlan, Mexico) and USA (Marfa, Texas). The number inside each parenthesis represents the number of PepMV isolates in that genotype/total number of samples submitted by growers for PepMV identification and genotype examination.
b nt: not tested.
The sequence identity of field collected PepMV isolates to those from commercial tomato seed lots
Analysis of the PepMV genetic diversity using the complete coat protein or partial RNA-dependant RNA polymerase (RdRP) genes showed the presence of quasispecies nature of PepMV in field populations throughout North America. Percentages of each sequence variants based on the coat protein gene were determined upon careful analysis to a multiple sequence alignment using 9 isolates from Canada and 27 isolates from the U.S. Despite such genetic diversity, the coat protein gene sequences among variants within the same genotype were very similar. There was only less than 1.0% sequence diversity among isolates in the PepMV CH2 genotype. A different gene (partial RdRP) was used to validate the genetic diversity of PepMV CH2 genotype as observed in 10 isolates from the U.S. [GenBank: JX866669 - JX866678]. Interestingly, the predominant CH2 variant (JX866670) shared an identical RdRP sequence to that of PepMV in a commercial tomato seed sample [‘F1Seed1’, VFTX12-32, GenBank: JX866669]. Among the 22 sequence variants from Texas, two major variant types were identified. Although the majority of CH2 variants from Texas (18 of 22) contained a typical coat protein gene (714 nt), 4 other sequence variants had an unique deletion of 12 nt (−GCTTCTAACCCA-) in the 5′ terminal portion of the coat protein gene between nt 19–30 resulting in a deletion of 4 aa (−ASNP-). Surprisingly, the same two major sequence variants were also identified in the commercial F1 hybrid tomato seed sample ‘F1Seed1’, including a normal coat protein gene [GenBank: JX866657] and a coat protein gene with 12 nt deletion [GenBank: KC579401]. A similar genetic diversity was also observed within the coat protein of 15 isolates in a different genotype (US1) from Mexico in 2012. Interestingly, a US1 isolate identified in another commercial tomato rootstock seed ‘RSSeed2’ [GenBank: JX8666635] was nearly identical to the 15 isolates from greenhouse tomatoes (GenBank: JX8666620-JX8666634). Such genetic diversity analysis strengthened the link between those of field epidemic isolates and those from commercial tomato seed lots, stressing the importance of planting PepMV-tested negative seed lots to reduce the chance of introducing contaminated planting materials into a production greenhouse.
Phylogenetic relationship and genome sequencing to the emerging genotypes
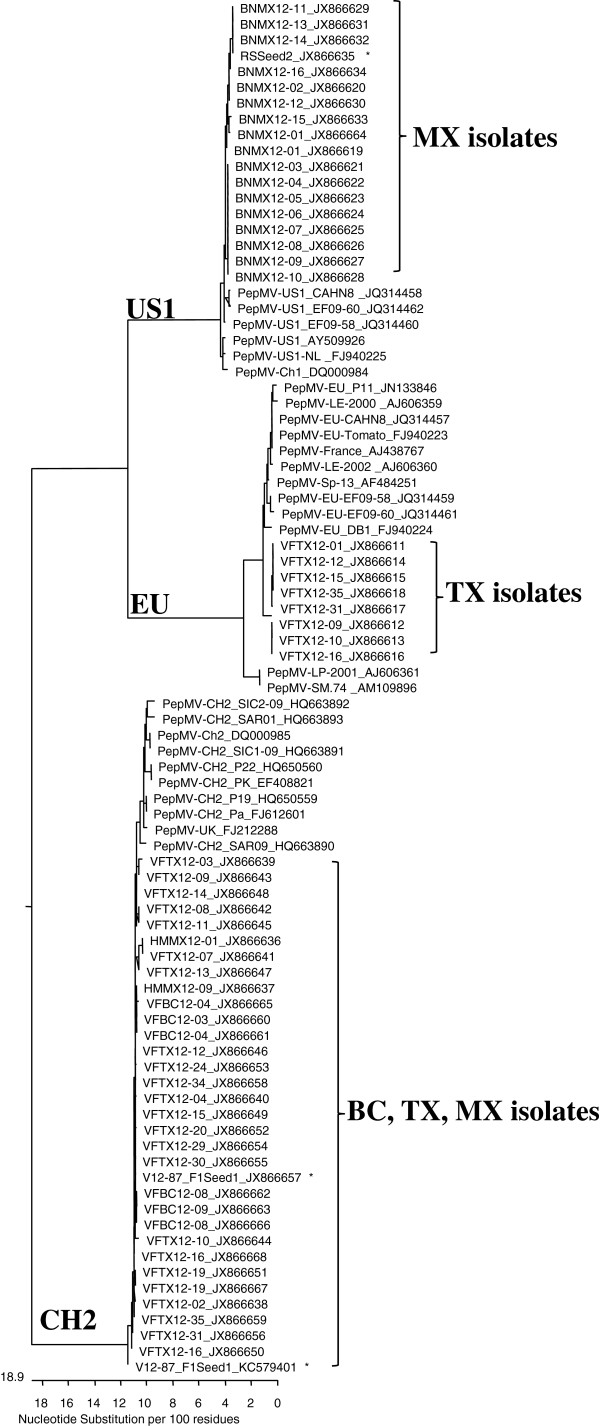
A phylogenetic tree was constructed by incorporating full coat protein genes from 53 PepMV isolates sequenced in the present study along with previously published PepMV sequences in GeneBank. As shown in Figure 2, all newly identified PepMV isolates could be assigned into three distinct phylogenetic lineages (or genotypes). The 16 Mexican isolates [with designations beginning with BNMX12-, GenBank: JX866619-JX866634] were grouped with the genotype US1. Eight U.S. isolates [with designations beginning with VFTX12-, GenBank: JX866611-JX866618] belonged to the genotype EU whereas another 28 isolates, including 22 from the U.S. [with designations beginning with VFTX12-, GenBank: JX866638-JX866659], four from Canada [with designations beginning with VFBC12-, GenBank: JX866660-JX866663), and two from a second location in Mexico [with designations beginning with HMMX12, GenBank: JX866636-JX866637), were found to belong to the CH2 genotype. All 10 Canadian isolates were assigned to the CH2 genotype but the U.S. isolates were separated in the CH2 and EU lineages. These CH2 variants from seed ‘F1Seed1’ [GenBank: JX866657 and KC579401] were clustered with other CH2 isolates from North American greenhouses. Although PepMV isolates from Mexico analyzed in the 2010 and 2011 growing seasons were genotyped as CH2 (Table 3), additional 16 isolates collected in 2012 from the same greenhouse belonged to genotype US1.
Figure 2.
Phylogenetic relationship of Pepino mosaic virus isolates analyzed using the coat protein gene sequences. The GenBank accession numbers for each virus isolate used were included in each entry. The 16 PepMV isolates collected from Mexico (MX) named as BNMX12- [GenBank: JX866619 – JX866634] clustered with the US1 lineage along with the isolate recovered from a commercial rootstock seed lot designated as ‘RSSeed2’, [GenBank: JX866635]. The two additional Mexican (MX) isolates in the CH2 lineage designated as HMMX12- [GenBank: JX866636 – JX866637] were collected from a different greenhouse in Mexico. The four Canadian (BC) isolates in the CH2 genotype are designated as VFBC12- [GenBank: JX866660 – JX866663]. The U.S. (TX) isolates shown as VFTX12-, including eight isolates in the EU genotype [GenBank: JX866611 – JX866618] and 22 representative isolates in the CH2 genotype [GenBank: JX866638 – JX866659], clustered along with two CH2 sequence variants recovered from a commercial tomato seed ‘F1Seed1’ [GenBank: JX866657 in a normal CP gene and KC579401 in a 12-nt deletion mutant]. For better identification, the three sequence variants obtained from the two seed samples were labeled with asterisks.
To gain better insights into these prevalent genotypes (CH2 and US1), extra efforts were taken to fully sequence the virus genomes in several selected isolates for comparison. These isolates included one US1 isolate from Mexico [BNMX12-01, GenBank: JX866664], two CH2 isolates from the U.S. [VFTX12-16, GenBank: JX866668; VFTX12-19, GenBank: JX866667] and two other CH2 isolates from Canada [VFBC12-04, GenBank: JX866665; VFBC12-08, GenBank: JX866666]. Sequence analysis demonstrated that the four CH2 isolates had 99.8% nucleotide sequence identity to one another but only 98.2% identity to the original Ch2 isolate [GenBank: DQ000985]. The two CH2 isolates from the U.S. (VFTX12-16 and VFTX12-19) with a 12-nt deletion in the coat protein gene were confirmed upon full genome sequencing.
Our previous surveys and genotyping efforts conducted in the 2006–2007 growing season showed a predominant EU genotype in Canada and the U.S. [17]. The CH2 genotype in the present study was consistently detected in grower’s samples from Canada and the U.S. since early 2010. Although the first natural PepMV infection was observed in 2010 in Mexico [18], the CH2 genotype was prevalent there for only two years until 2011 (Table 3), before being displaced by the genotype US1 in 2012 (Figure 2).
Although seed transmission of PepMV in tomato may occur sporadically due to the virus being localized on the seed coat (testa) and not in the embryo [32], tomato seedling infection can occur either through seedling germination [33] or by mechanical inoculation [32]. However, how these PepMV CH2 isolates were initially introduced into European greenhouse tomatoes was not clear [14,29-31]. It is possible that these CH2 isolates could be introduced in the same manner as those described here, because many European greenhouse tomato growers use similar sources of commercial tomato seeds as American growers. Moreover, sequences from these European CH2 isolates are very similar to those from North America as indicated in the phylogenetic analysis (Figure 2). The Chilean isolates (Ch1 = US1 genotype, Ch2 = CH2 genotype) were identified in a commercial seed lot imported from Chile in 2004 [27]. A CH2 isolate was also detected in 2007 in Texas [17]. In just a few years, the CH2 genotype replaced the EU genotype and emerged as a predominant strain in causing pepino mosaic disease epidemics in many greenhouse tomato facilities around the world. Gomez and colleagues [30] demonstrated a higher genetic fitness for a CH2 isolate in a mixed infection of an EU isolate. The data from the current study appeared to support such notion in that the titer of EU in the three Texan samples (VFTX12-01 to VFTX12-03) mixed infected with CH2 was so low that it was not detectable by RT-PCR. Only after the use of more sensitive RT-LAMP was the presence of EU sequence confirmed (Table 2). Similarly, in three Canadian samples (VFBC12-01 to VFBC12-03) containing a mixed infection of CH2 and US1, the presence of US1 sequence was detected only by the use of RT-LAMP (Table 2). Such genotype displacement could also be enhanced by several environmental and ecological factors associated with intense greenhouse tomato production system. In addition, the human factor in performing intensive hands-on operations can play an important role in PepMV transmission once introduced in a greenhouse facility. Despite the increasing demand and requirement in seed health test for PepMV, the current standard seed health test as recommended by International Seed Health Initiative (ISHI) with ELISA test may not be sensitive enough to detect a low level of seed contamination in commercial seed lots. In two seed samples that were initially negative for PepMV in ELISA, the virus was revealed by immunocapture real-time RT-PCR [48]. Although the Ct values were close to the upper end of the “positive” cutoff (Ct = 35), with Ct = 33.67 for ‘F1Seed1’ and Ct = 32.24 for ‘RSSeed2’while the PepMV-positive control seed had a Ct = 25.53. Subsequently, positive identification was confirmed using the genotype-specific RT-PCR system through direct sequencing and sequence analysis of the amplified RT-PCR product.
The three major genotypes of PepMV (EU, US1 and CH2) have been recognized in North America [16,17,21]. However, we could not confirm the presence of US2 genotype either through Sanger sequencing or by small RNA deep sequencing [49]. The original US2 genotype sequence [16] was probably either a natural recombinant or an artifact between the two recognized genotypes (US1 and EU), and thus was not considered further as one of the representative genotypes for PepMV. We also did not detect the so-called LP genotype [12] in our surveys of PepMV on commercial greenhouse tomatoes in North America.
The LAMP technology is highly specific for the target sequence and has been developed as a powerful tool for viral disease diagnosis [39-47]. In the present study, we were interested in developing genotype –specific RT-LAMP that would permit an effective and efficient genotyping without lengthy sequencing. Initial genotype analysis was performed using sequencing of RT-PCR products containing a small portion of the viral genome [17]. This sequencing method is accurate, but is laborious and time consuming. To allow for an efficient assessment of many field-collected samples, it was necessary to develop and apply a simple technology. The genotype-specific RT-LAMP developed in the current study was effective for PepMV genotyping and its specificity was validated using RT-PCR and sequencing. Although the amplified RT-LAMP products can be detected in a number of ways, in the current study, we chose a direct staining with SYBR Green in an enclosed tube to minimize potential cross-contamination. Thus, the genotype-specific RT-LAMP technology could be useful in field setting to analyze PepMV isolates for their genetic composition.
The highly contagious nature of PepMV makes it extremely difficult to control. Cross-protection has been shown to be an effective measure only if it is used against a virulent isolate belonging to the same genotype [50]. Cross-protection is not effective against a PepMV isolate in a different genotype and could potentially result in even more serious disease in a mixed infection [15]. Therefore, understanding the genetic diversity is critical in deciding which mild strain of the virus should be used to achieve a successful cross protection.
Conclusions
The population genetic analysis in the present study showed a shift in the prevalent genotype of PepMV from the EU to the CH2 genotype since 2010 in North America. This result is in agreement with the trend observed in several European countries [14,29-31]. In the present study, we revealed another genotype shifting event from CH2 to US1 in Mexico only two years after the first identification of PepMV infection there [18]. Such genotype shift was likely not induced through a natural virus mutation in such a short period of time but rather from an unintentional introduction of a more aggressive genotype of PepMV through the use of contaminated tomato seed lots. This hypothesis was supported by the high sequence identity between the PepMV isolates originated from greenhouse tomatoes and those from commercial tomato seed sources (scion and rootstock). Taken together, our genetic diversity characterization and phylogenetic analysis supported the notion that contaminated tomato seed lots were the sources of initial inocula for disease epidemic, which resulted in a dramatic genotype shifting in North America from one year to another. The application of genotype-specific RT-LAMP can enhance our capacity to monitor field genotype composition and to apply appropriate strategies to manage pepino mosaic disease.
Materials and methods
Survey, culture collection and detection of PepMV
In collaboration with three major greenhouse tomato growers in North America, we initiated a surveillance system to monitor the occurrence of PepMV in Canada, Mexico and the United States and to characterize the genetic diversity of field populations of PepMV. After collection, samples were shipped to our lab at the U.S. Vegetable Laboratory (Charleston, SC, USA) using appropriate plant pathogen diagnostic permits. Upon receipt, samples were inspected for symptom expression and tested for the presence of PepMV by enzyme-linked immunosorbent assay (ELISA) (Agdia, USA), AgriStrip (BioReba, Switzerland), or immunocapture real-time RT-PCR [48]. Occasionally, virus infectivity was assayed by mechanical inoculation of tomato or Nicotiana benthamiana plants [32]. The collected tissue samples were at different stages of plant development with symptoms ranging from foliar mosaic to marbling on fruits. Thus, the virus isolates reported in this study were representative of a broad spectrum of PepMV populations in these greenhouses. To characterize genetic diversity of PepMV and to monitor a dynamic of genotype shift over the years, tomato samples collected and tested positive for PepMV by ELISA from the same representative greenhouse in each of the three countries in North America were used for molecular characterization. In the present study, fourteen samples originated from Canada, four collected in 2010 and ten in 2012. Twenty four samples were collected from Mexico, three in 2010, five in 2011, and sixteen in 2012. Forty samples were from the United States, including four in 2009, three in 2010, one in 2011, and thirty two in 2012. In addition, two commercial tomato seed samples (designated as ‘F1Seed1’ for a F1 hybrid and ‘RSSeed2’ for a rootstock) were a gift from an anonymous donor. According to the International Seed Health Initiative standard method for PepMV [51], seed health tests were performed with 3,000 seeds divided into 12 subsamples of 250 seeds each using ELISA (Agdia, USA) or immunocapture real-time RT-PCR [48].
Development of genotype specific RT-PCR
Initial genotype analysis for PepMV was based on sequencing the amplified RT-PCR products using two sets of primers targeting two genomic regions to allow for accurate genotyping [17]. The first generic primer pair KL05-13 (5′ GTC CTC ACC AAT AAA TTT AG 3′) and KL05-14 (5′ AGG AAA ACT TAA CCC GTT C 3′) generated a relatively short genomic sequence (202 bp) targeting the TGB2-3 region which was useful for general PepMV genotyping among EU, US1 and CH2. The second primer set KL04-52 (5′ GTG CTT ACA GTT CTG ACA TC 3′) and KL05-20 (5′ CTA GAA TTG GCA CTT TGC AC 3′) targeting the CH2 genome in the RNA dependant RNA polymerase (RdRp) region were used to confirm the identification of a CH2 isolate [17]. Therefore, this process was cumbersome and time consuming. To allow for more efficient genotyping, three genotype-specific primer sets were designed based on the conserved sequence region in each genotype in this study (Table 1). Using these primer sets, RT-PCR amplified products contained sequences of the coat protein gene and a portion of 3′ untranslated region for each specific genotype. To facilitate direct sequencing of the RT-PCR products, primers were designed to contain the M13 forward or reverse primer sequences. RT-PCR was conducted using a template viral RNA from immunocapture [48], or with a total plant RNA prepared using an RNeasy plant kit (Qiagen, USA) or a TRIzol reagent (Invitrogen, USA). RT-PCR was conducted using a Superscript III one-Step RT-PCR kit with RT/Platinum Taq Hifi enzyme mix (Invitrogen, USA). The cycling parameters included a reverse transcription for 30 min at 50°C and a denaturation for 2 min at 94°C, followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 1 min, with a final extension cycle of 10 min at 72°C. An aliquot of RT-PCR preparation (10 μl) from each reaction was applied onto a 1.5% agarose gel for electrophoresis and the RT-PCR products were visualized under a UV transilluminator after staining with 0.01% SYBR Safe (Invitrogen, USA). Gel images were taken using a ChemiDoc digital imaging system (Bio-Rad, USA). To confirm the genotype specificity, nucleotide sequencing was conducted. Upon purification using a QIAquick PCR Purification kit (Qiagen, USA), RT-PCR products were used for direct sequencing or cloned into a pCR4-TOPO vector (Invitrogen, USA). Sanger sequencing was performed by Functional Genomics (Madison, WI, USA). Sequence assembly and alignment analysis were performed with DNASTAR Lasergene 10 (Madison, WI, USA). In addition, complete genomic sequences from selected isolates representing CH2 or US1 were determined using overlapping RT-PCR products generated with genotype specific primers for CH2 (Table 1) or for US1 [49].
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) for genotype-specific detection
Through multiple alignments of complete genomic PepMV sequences, a conserved region (~99%) among isolates within each particular genotype (EU, US1, or CH2) was identified. Using consensus sequences from the identified conserved region (about 200 nt), a set of primers suitable for RT-LAMP was designed using PrimerExplorer V4 software [52] and synthesized by Sigma Genosys (USA) (Table 1). The RT-LAMP assay was conducted using a RNA amplification kit from Eiken Chemical (Japan) following the manufacturer’s instructions. Each 25 μl reaction consisted of 12.5 μl of 2× reaction buffer [containing 40 mM Tris–HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6M Betaine, and 2.8 mM each of dNTPs], 2 μl of 20 μM forward inner primer (FIP), 2μl of 20 μM back inner primer (BIP), 1 μl of 20 μM of Loop-F primer, 1μl of 20 μM of Loop-B primer, 0.25 μl of 20 μM of F3 primer, 0.25 μl of 20 μM of B3 primer, 1.0 μl of enzyme mix (reverse transcriptase and DNA polymerase) and 2.0 μl of RNA preparation. The mixture was incubated at 65°C for 60 min, followed by a denaturation for 2 min at 95°C to terminate the reaction. Products were checked by electrophoresis on a 2% agarose gel containing 1:10,000 diluted SYBR gel stain or directly visualized by addition of 1.0 μl of 1:10 diluted SYBR Green I (Invitrogen, USA). In some cases, to validate the sequence specificity, RT-LAMP products were cloned with a TOPO TA cloning kit (Invitrogen, USA) after tailing with dATP in 1x GoTag buffer (Promega, USA) at 72C for 15 minutes. Upon screening, colonies with inserts were selected, sequenced and analyzed using BLASTn in the National Center for Biotechnology Information (NCBI) database.
Phylogenetic analysis
To assess genetic composition of PepMV populations in samples collected from North America, the coat protein gene region were aligned and analyzed with CLUSTAL-W [53]. A phylogenetic relationship for these PepMV isolates was assessed using the neighbor-joining method [54] in DNASTAR Lasergene 10 in 1,000 bootstrap iterations to generate a robust phylogenetic tree.
Abbreviations
PepMV: Pepino mosaic virus; ELISA: Enzyme-linked immunosorbent assay; RT-PCR: Reverse transcription polymerase chain reaction; RT-LAMP: Reverse transcription loop-mediated isothermal amplification; ISHI: International Seed Health Initiative; RdRP: RNA-dependant RNA polymerase; NCBI: National Center for Biotechnology Information.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
KSL conceived the study, designed methods and experiments; performed experiments and sequence analysis; analyzed data and interpretation, drafted and revised the manuscript. RL designed primers for RT-LAMP; performed RT-LAMP; conducted sequence analysis; evaluated data and revised the manuscript. MB coordinated activities in greenhouse survey, sample collection, and revised manuscript. All the authors have read and approved the final manuscript.
Contributor Information
Kai-Shu Ling, Email: kai.ling@ars.usda.gov.
Rugang Li, Email: Rugang.li@ars.usda.gov.
Michael Bledsoe, Email: mbledsoe@villagefarms.com.
Acknowledgements
We thank Dr. W. Patrick Wechter for critical reading of the manuscript and Andrea Gilliard and Emily Fillippeli for their excellent technical assistance. The field survey and sample collection efforts from participating growers in Canada, Mexico and the U.S. are greatly appreciated. This work is supported in-part by a grant from the USDA, National Institute of Food and Agriculture, Specialty Crop Research Initiative Program (SCRI-600-25320) to KSL. Funding agency had no role in experiment design, data collection, analysis or interpretation, nor in manuscript writing, revision or in manuscript submission.
References
- FAOSTAT. [ http://faostat.fao.org/] (last accessed March 27, 2013)
- Cook RL, Calvin L. Economic Research Report. 2. Washington, DC: Economic Research Service, U.S. Department of Agriculture; 2005. Greenhouse tomatoes change the dynamics of the North American fresh tomato industry; pp. 1–81. [Google Scholar]
- Hanssen IM, Lapidot M, Thomma BPHJ. Emerging viral diseases of tomato crops. Mol Plant-Microbe Interact. 2010;23:539–548. doi: 10.1094/MPMI-23-5-0539. [DOI] [PubMed] [Google Scholar]
- Jones RAC, Koenig R, Lesemann DE. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricantum) Ann Appl Biol. 1980;94:61–68. doi: 10.1111/j.1744-7348.1980.tb03896.x. [DOI] [Google Scholar]
- van der Vlugt RAA, Stijger CCMM, Verhoeven JTJ, Lesemann DE. First report of Pepino mosaic virus on tomato. Plant Dis. 2000;84:103. doi: 10.1094/PDIS.2000.84.1.103C. [DOI] [PubMed] [Google Scholar]
- Mumford RA, Metcalfe EJ. The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch Virol. 2001;146:2455–2460. doi: 10.1007/s007050170015. [DOI] [PubMed] [Google Scholar]
- Jorda C, Lazaro-Perez A, Martinez-Culebras PV, Abad P, Lacasa A, Guerrero MM. First report of Pepino mosaic virus on tomato in Spain. Plant Dis. 2001;85:1292. doi: 10.1094/PDIS.2001.85.12.1292C. [DOI] [PubMed] [Google Scholar]
- Roggero P, Masenga V, Lenzi R, Coghe F, Ena S, Winter S. First report of Pepino mosaic virus in tomato in Italy. Plant Pathol. 2001;50:798–800. doi: 10.1046/j.1365-3059.2001.00621.x. [DOI] [Google Scholar]
- Aguilar JM, Hernandez-Gallarod MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol. 2002;147:2009–2015. doi: 10.1007/s00705-002-0848-9. [DOI] [PubMed] [Google Scholar]
- Cotillon AC, Girard M, Ducouret S. Complete nucleotide sequence of the genomic RNA of a French isolate of Pepino mosaic virus (PepMV) Arch Virol. 2002;147:2231–2238. doi: 10.1007/s00705-002-0873-8. [DOI] [PubMed] [Google Scholar]
- Verhoeven JTJ, van der Vlugt RAA, Roenhorst JW. High similarity between tomato isolates of Pepino mosaic virus suggests a common origin. Eur J Plant Pathol. 2003;109:419–425. doi: 10.1023/A:1024261121468. [DOI] [Google Scholar]
- Lopez C, Soler S, Nuez F. Comparison of the complete sequences of three different isolates of Pepino mosaic virus: size variability of the TGBp3 protein between tomato and L. peruvianum isolates. Arch Virol. 2005;150:619–627. doi: 10.1007/s00705-004-0438-0. [DOI] [PubMed] [Google Scholar]
- Pospieszny H, Borodynko N. New Polish isolate of Pepino mosaic virus highly distinct from European tomato, Peruvian, and US2 strains. Plant Dis. 2006;90:1106–1108. doi: 10.1094/PD-90-1106C. [DOI] [PubMed] [Google Scholar]
- Hanssen IM, Paeleman A, Wittemans L, Goen K, Lievens B, Bragard C, Vanachter ACRC, Thomma BPHJ. Genetic characterization of Pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol. 2008;121:131–146. doi: 10.1007/s10658-007-9255-0. [DOI] [Google Scholar]
- French CJ, Bouthillier M, Bernardy M, Ferguson G, Sabourin M, Johnson R, Masters C, Godkin S, Mumford R. First report of Pepino mosaic virus in Canada and the United States. Plant Dis. 2001;85:1121–1123. doi: 10.1094/PDIS.2001.85.10.1121B. [DOI] [PubMed] [Google Scholar]
- Maroon-Lango CJ, Guaragna MA, Jordan RL, Hammond J, Bandla M, Marquardt SK. Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch Virol. 2005;150:1187–1201. doi: 10.1007/s00705-005-0495-z. [DOI] [PubMed] [Google Scholar]
- Ling K-S, Wintermantel WM, Bledsoe M. Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis. 2008;92:1683–1688. doi: 10.1094/PDIS-92-12-1683. [DOI] [PubMed] [Google Scholar]
- Ling K-S, Zhang W. First report of Pepino mosaic virus infecting tomato in Mexico. Plant Dis. 2011;95:1035. doi: 10.1094/PDIS-04-11-0334. [DOI] [PubMed] [Google Scholar]
- Soler S, Prohens J, Diez MJ, Nuez F. Natural occurrence of Pepino mosaic virus in Lycopersicon species in central and southern Peru. J Phytopathol. 2002;150:49–53. doi: 10.1046/j.1439-0434.2002.00712.x. [DOI] [Google Scholar]
- Soler S, Lopez C, Nuez F. Natural occurrence of viruses in Lycopersicon spp. in Ecuador. Plant Dis. 2005;89:1244–1254. doi: 10.1094/PD-89-1244C. [DOI] [PubMed] [Google Scholar]
- Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes. 2007;34:1–8. doi: 10.1007/s11262-006-0003-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shen ZJ, Zhong J, Lu XL, Cheng G, Li RD. Preliminary characterization of Pepino mosaic virus Shanghai isolate (PepMV-Sh) and its detection with ELISA. Acta Agric Shanghai. 2003;19:90–92. [Google Scholar]
- Fakhro A, von Bargen S, Bandte M, Büttner C. Pepino mosaic virus, a first report of a virus infecting tomato in Syria. Phytopathol Mediterr. 2010;49:99–101. [Google Scholar]
- Carmichael DJ, Rey MEC, Naidoo S, Cook G, van Heerden SW. First report of Pepino mosaic virus infecting tomato in South Africa. Plant Dis. 2011;95:767. doi: 10.1094/PDIS-01-11-0036. [DOI] [PubMed] [Google Scholar]
- van der Vlugt RAA. Pepino mosaic virus. Hellenic Plant Prot J. 2009;2:47–56. [Google Scholar]
- Hanssen IM, Thomma BPHJ. Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol Plant Pathol. 2010;11:179–189. doi: 10.1111/j.1364-3703.2009.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence NJ, Basham J, Mumford RA, Hayman G, Edmondson R, Jones DR. Effect of Pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the United Kingdom. Plant Pathol. 2006;55:595–606. doi: 10.1111/j.1365-3059.2006.01406.x. [DOI] [Google Scholar]
- Pagan I, Cordoba-Selles M, Martinez-Priego L, Fraile A, Malpica J, Jorda C, Garcia-Arenal F. Genetic structure of the population of Pepino mosaic virus infecting tomato crops in Spain. Phytopathology. 2006;96:274–279. doi: 10.1094/PHYTO-96-0274. [DOI] [PubMed] [Google Scholar]
- Pospieszny H, Hasiów B, Borodynko N. Characterization of two distinct Polish isolates of Pepino mosaic virus. Eur J Plant Pathol. 2008;122:443–445. doi: 10.1007/s10658-008-9280-7. [DOI] [Google Scholar]
- Gomez P, Sempere RN, Elena SF, Aranda MA. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol. 2009;83:12378–12387. doi: 10.1128/JVI.01486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasiow-Jaroszewska B, Jackowiak P, Borodynko N, Figlerowicz M, Pospieszny H. Quasispecies nature of Pepino mosaic virus and its evolutionary dynamics. Virus Genes. 2010;41:260–267. doi: 10.1007/s11262-010-0497-0. [DOI] [PubMed] [Google Scholar]
- Ling K-S. Pepino mosaic virus on tomato seed: virus location and mechanical transmission. Plant Dis. 2008;92:1701–1705. doi: 10.1094/PDIS-92-12-1701. [DOI] [PubMed] [Google Scholar]
- Hanssen IM, Mumford R, Blystad DR, Cortez I, Hasiów-Jaroszewska B, Hristova D, Pagán I, Pereira A-M, Peters J, Pospieszny H, Ravnikar M, Stijger I, Tomassoli L, Varveri C, van der Vlugt R, Nielsen SL. Seed transmission of Pepino mosaic virus in tomato. Eur J Plant Pathol. 2010;126:145–152. doi: 10.1007/s10658-009-9528-x. [DOI] [Google Scholar]
- Alfaro-Fernandez A, Sánchez-Navarro JA, Cebrián MC, Córdoba-Sellés MC, Pallás V, Jordá C. Simultaneous detection and identification of Pepino mosaic virus (PepMV) isolates by multiplex one-step RT-PCR. Eur J Plant Pathol. 2009;125:143–158. doi: 10.1007/s10658-009-9466-7. [DOI] [Google Scholar]
- Martinez-Culebrae PV, Lazaro A, Abad Campos P, Jorda C. A RT-PCR assay combined with RFLP analysis for detection and differentiation of isolates of Pepino mosaic virus (PepMV) from tomato. Eur J Plant Pathol. 2002;108:887–892. doi: 10.1023/A:1021247220932. [DOI] [Google Scholar]
- Gutiérrez-Aguirre I, Mehle N, Delić D, Gruden K, Mumford R, Ravnikar M. Real-time quantitative PCR based sensitive detection and genotype discrimination of Pepino mosaic virus. J Virol Methods. 2009;162:46–55. doi: 10.1016/j.jviromet.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Boubourakas IN, Fukuta S, Kyriakopoulou PE. Sensitive and rapid detection of Peach latent mosaic viroid by the reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2009;160:63–68. doi: 10.1016/j.jviromet.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Fukuta S, Kato S, Yoshida K, Mizukami Y, Ishida A, Ureda J, Kanbe M, Ishimoto Y. Detection of Tomato yellow leaf curl virus by loop-mediated isothermal amplification reaction. J Virol Methods. 2003;112:35–40. doi: 10.1016/S0166-0934(03)00187-3. [DOI] [PubMed] [Google Scholar]
- Fukuta S, Ohishi K, Yoshida K, Mizukami Y, Ishida A, Kanbe M. Development of immunocapture reverse transcription loop-mediated isothermal amplification for the detection of Tomato spotted wilt virus from chrysanthemum. J Virol Methods. 2004;121:49–55. doi: 10.1016/j.jviromet.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Fukuta S, Takeyama K, Suzuki M, Shichi A, Ichikawa K, Nakanishi H. Detection of Kyuri green mottle mosaic virus from soil by the immunocapture reverse transcription loop-mediated isothermal amplification reaction. Plant Pathol J. 2012;11:51–59. doi: 10.3923/ppj.2012.51.59. [DOI] [Google Scholar]
- Hadersdorfer J, Neumuller M, Treutter D, Fischer TC. Fast and reliable detection of Plum pox virus in woody host plants using the Blue LAMP protocol. Ann Appl Biol. 2011;159:456–466. doi: 10.1111/j.1744-7348.2011.00510.x. [DOI] [Google Scholar]
- Le DT, Netsu O, Uehara-Ichiki T, Shimizu T, Choi IR, Omura T, Sasaya T. Molecular detection of nine rice viruses by a reverse-transcription loop-mediated isothermal amplification assay. J Virol Methods. 2010;170:90–93. doi: 10.1016/j.jviromet.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Nie X. Reverse transcription loop-mediated isothermal amplification of DNA for detection of Potato virus Y. Plant Dis. 2005;89:605–610. doi: 10.1094/PD-89-0605. [DOI] [PubMed] [Google Scholar]
- Tsutsumi N, Yanagisawa H, Fujiwara Y, Ohara T. Detection of Potato spindle tuber viroid by reverse transcription loop-mediated isothermal amplification. Bull Pl Prot Japan. 2010;46:61–67. [Google Scholar]
- Zhang Z-Y, Liu X-J, Li DW, Yu J–L, Han C-G. Rapid detection of Wheat yellow mosaic virus by reverse transcription loop-mediated isothermal amplification. Virol J. 2011;8:550. doi: 10.1186/1743-422X-8-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KS, Wechter WP, Jordan R. Development of a one-step immunocapture real-time TaqMan RT-PCR assay for the broad spectrum detection of Pepino mosaic virus. J Virol Methods. 2007;144:65–72. doi: 10.1016/j.jviromet.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Li R, Gao S, Hernandez AG, Wechter WP, Fei Z, Ling K-S. Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS ONE. 2012;7:e37127. doi: 10.1371/journal.pone.0037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen IM, Gutie´rrez-Aguirre I, Paeleman A, Goen K, Wittemans L, Lievens B, Vanachter ACRC, Ravnikar M, Thomma BPHJ. Cross-protection or enhanced symptom display in greenhouse tomato co-infected with different Pepino mosaic virus isolates. Plant Pathol. 2010;59:13–21. doi: 10.1111/j.1365-3059.2009.02190.x. [DOI] [Google Scholar]
- International seed health initiative. [ http://www.worldseed.org/isf/ishi_vegetable.html] (last accessed March 27, 2013)
- LAMP primerExplorer. [ http://primerexplorer.jp/elamp4.0.0/index.html] (last accessed March 27, 2013)
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-joining Method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]