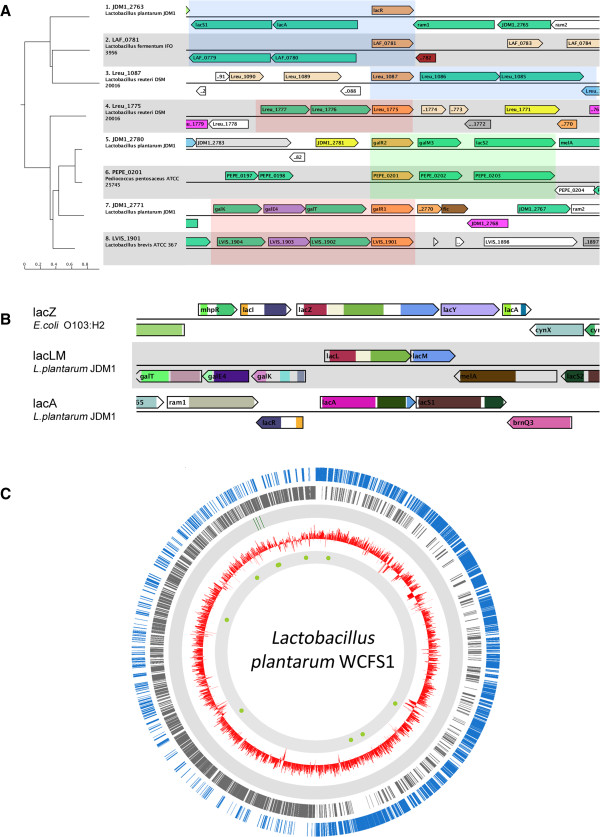
Figure 2.
EbgR-like transcription factors in L. plantarum and other lactobacilli. A) MGcV visualization of a phylogenetic tree of EbgR-type regulators in some Lactobacilli. To simplify, the tree was pruned (species: Lactobacillus plantarum JDM1 and WCFS1, Lactobacillus fermentum IFO3956, Lactobacillus reuteri DSM20016, Lactobacillus brevis ATCC 367 and Pediococcus pentosaceus). The context range was set to 10.000 nucleotides, genes were colored by COG-class and trivial names were used to label genes. The visual combination of the phylogenetic tree and genomic context allows to distinguish three groups; lacR-, rafR- and galR-like sequences (designated in blue, green and red, respectively). B) Comparative context map of the beta-galactosidase encoding genes lacZ (E. coli), lacLM (L. plantarum) and lacA (L. plantarum). Pfam domains are used for gene-coloring and trivial names are used to label genes. In L. plantarum lacLM and lacA are both annotated to encode a protein with the same name (beta-galactosidase). Yet, from this comparative view it becomes clear that lacLM and lacA must have a different evolutionary origin. Although lacLM is encoded by two genes, the domain structure appears identical to the single lacZ gene of E. coli. C) A circular genome map of L. plantarum in which the ORFs on the plus strand (blue), on the minus stand (grey), the locations of regulator encoding genes lacR, rafR and galR (green), the GC% (red) and putative binding sites (similarity to motif >90% [8]; represented by the green dots) are included.