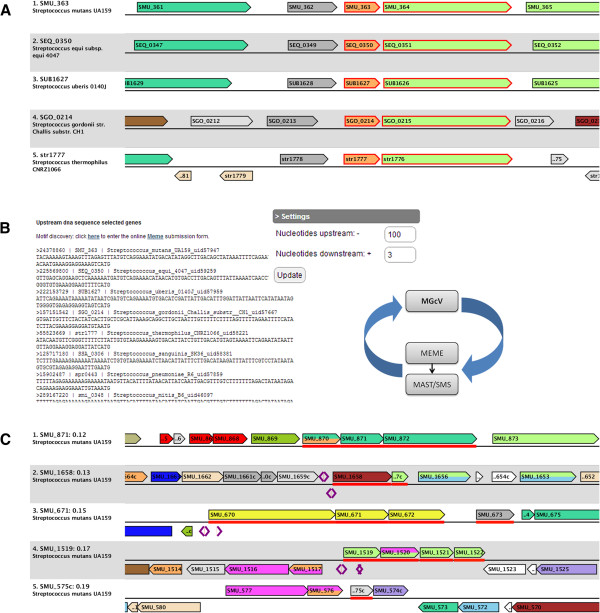
Figure 3.
Application of MGcV in the reconstruction of GlnR-mediated regulation in Streptococcus mutans. A) Comparative context map of GlnR homologs obtained via a BLAST search in all sequenced Streptococci (only five species are shown). The GlnRA operon and its direct genomic context is clearly conserved in the Streptococci. The map was used to graphically select those genes that could be preceded by a binding site. B) The “upstream region” option of the “Data-export”-box was used to obtain upstream regions of the selected genes. Subsequently, the available link to MEME was used to search for possible overrepresented motifs. The results were refined and a motif defined [63], which was then used to search and score putative binding sites (e.g. using MAST or SMS). C) A comparative context map ranked on expression ratios (low-to-high) of a GlnR mutant, visualized in conjunction with predicted GlnR binding sites. To exemplify, the figure is limited to the top 5 of down-regulated genes. In this map, gene expression ratios are represented in a colored bar (red-to-green gradient; red is down-regulated) at the baseline and putative binding sites are designated by purple arrows (direction representing the strand). Both the microarray data and putative binding sites and their corresponding binding sites were uploaded using the “Data import”-box. The resulting map allows the analysis of the putative GlnR binding sites in light of the expression data of the GlnR mutant. Most of the top down regulated genes (SMU_1658, SMU_671 and SMU,1519 in Figure 3C) indeed are preceded by a putative binding site.