Abstract
Recombination during meiosis in the form of crossover events promotes the segregation of homologous chromosomes by providing the only physical linkage between these chromosomes. Recombination occurs not only between allelic sites but also between non-allelic (ectopic) sites. Ectopic recombination is often suppressed to prevent non-productive linkages. In this study, we examined the effects of various mutations in genes involved in meiotic recombination on ectopic recombination during meiosis. RAD24, a DNA damage checkpoint clamp-loader gene, suppressed ectopic recombination in wild type in the same pathway as RAD51. In the absence of RAD24, a meiosis-specific recA homolog, DMC1, suppressed the recombination. Homology search and strand exchange in ectopic recombination occurred when either the RAD51 or the DMC1 recA homolog was absent, but was promoted by RAD52. Unexpectedly, the zip1 mutant, which is defective in chromosome synapsis, showed a decrease, rather than an increase, in ectopic recombination. Our results provide evidence for two types of ectopic recombination: one that occurs in wild-type cells and a second that occurs predominantly when the checkpoint pathway is inactivated.
Introduction
Meiotic recombination ensures the segregation of homologous chromosomes at meiosis I by providing a physical linkage between them. Although this recombination generates both crossover (CO) and non-crossover (NCO) recombination products, only COs generate physical linkages between homologs known as chiasmata [1], [2]. Recombination during meiosis occurs predominantly between homologous chromosomes (inter-homolog recombination bias), although it has been reported that recombination between sister chromatids also occurs at reasonable frequency [3], [4]. On the other hand, during mitosis, recombination preferentially takes place between sister chromatids [5]. In addition, in some cases, recombination also promotes exchange between non-allelic (ectopic) sites on chromosomes. This non-allelic homologous recombination (NAHR) may result in chromosome translocations, deletions or inversions, which have been associated with instability of the genome [4].
Meiotic CO formation is initiated by the generation of double-strand breaks (DSBs) [6]. After nucleolytic processing of DSB ends, exposed single-stranded DNA is used by the recombination machinery to search for corresponding DNA sequences on a homologous chromosome (as opposed to a sister chromatid). One end of each DSB site is believed to engage in direct interaction with homologous sequences, whereas the other end does not participate in the initial homology search but is involved later in a step called “second-end capture” [7]–[9]. After identifying the complementary DNA sequence on the homolog, single-stranded DNA is thought to form an unstable D-loop. If a DNA strand synthesized from the invading 3′-end is dissociated from the template strand in the D-loop, it leads to the formation of NCO products through a synthesis-dependent strand-annealing pathway [7], [10], [11]. Alternatively, if the synthesized strand is not dissociated, the D-loop may be converted into a “stable” single-stranded invasion intermediate (SEI) [7]. Further processing leads to the formation of intermediates with two Holliday junctions, referred to as a double-Holliday junction (dHJ) [12], that are specifically resolved into reciprocal CO products [7], [10], [13].
In meiotic recombination, two RecA homologs, Rad51 and Dmc1, play a critical role in the homology search process [14], [15]. Dmc1 strand exchange activity is sufficient for catalyzing D-loop formation during meiosis, although the presence of the Rad51 protein is necessary to regulate the activity of Dmc1 [16]–[19]. Coordinated action of the two RecA homologs is necessary for inter-homolog recombination bias [18], [19]. The assembly of Rad51 depends on Rad52, the Rad55-Rad57 complex and PCSS (Psy3-Csm2-Shu1-Shu2)/Shu complex as well as the single-stranded DNA binding protein RPA (Replication protein-A) [17], [20]–[22], whereas Dmc1 assembly depends on Mei5-Sae3 and Rad51 [19], [23]-[26]. Strand invasion by Rad51 and Dmc1 is facilitated by two Swi2/Snf2 DNA translocases, Rad54 and Tid1/Rdh54 [27]–[29].
Meiotic CO formation is also facilitated by a group of proteins called ZMM (Zip, Mer, Msh) or SIC (synaptic initiation complex), hereafter referred to as ZMM [13], [30]–[32]. ZMM proteins include Zip1, Zip2, Zip3, Mer3, Msh4, Msh5, Spo22/Zip4 and Spo16. Among these, Msh4 and Msh5 are MutS homologs [33]–[35]. The assembly of ZMM foci depends on DSB formation and processing [30]–[32], [36]. ZMM proteins also coordinate the formation of the synaptonemal complex (SC), a ladder-like structure consisting of two chromosomal axes flanking a central region, with the recombination. Zip1 is a component of the central region of the SC [37], [38]. Although, in the yeast, recombination promotes chromosome synapsis, chromosome synapsis is thought to control the processing of recombination intermediates between homologous chromosomes [39].
During meiosis, DNA damage checkpoint proteins play an important role in the cellular response to DNA damage as well as in DNA repair [40]. In the mitotic DNA damage checkpoint pathway of Saccharomyces cerevisiae, the Rad24-RFC (Rad17-RFC in other organisms) clamp-loader complex promotes the assembly of the Ddc1-Rad17-Mec3 clamp at the junction of tailed DNAs and activates downstream events [41], [42]. The Ddc1-Rad17-Mec3 clamp is referred as to the “9-1-1” (Rad9-Rad1-Hus1) complex in other organisms. In parallel, Mec1/ATR kinase is recruited to and activated on RPA-coated single-stranded DNAs in a Ddc2/ATRIP-dependent manner [43]. In meiosis, as in mitosis, checkpoint protein loading at the sites of recombination intermediates can induce delay in the entry into meiosis I [44]. In addition to providing a means to block meiotic progression when recombination is incomplete, the checkpoint proteins work directly in meiotic recombination [45]–[47]. For example, checkpoint proteins such as Rad17, Rad24 and Mec1 restrict the use of sister chromatids or repeated sequences at non-allelic (ectopic) sites as recombination partners [45], [47]. A direct role for checkpoint proteins in the recombination process is also suggested by the reduced rate of recombination in mutants lacking checkpoint function [46]. Consistent with this, over-expression of RAD51 or RAD54 can suppress the meiotic defects of checkpoint mutants as well as mitotic DNA damage sensitivity [46]. Moreover, the checkpoint clamp loader and clamp promotes CO formation by recruiting ZMM proteins to the chromosomes (Our unpublished results).
Previous genetic analyses showed that ectopic recombination occurs at frequencies similar to allelic recombination [48]–[52]. These studies showed that ectopic recombination between closely linked loci occurs more frequently than between dispersed loci. [48], [50], [51]. However, a genetic pathway(s) defining meiotic ectopic recombination (or NAHR) largely remains unknown. Genetic analysis, which requires viable gametes, is not applicable to mutants with reduced spore viabilities. The development of a physical assay for monitoring ectopic recombination has made it possible to dissect the pathway(s) that create these events and has revealed a critical role for DNA damage checkpoint genes in this process [45]. In this paper, we describe relationships among RAD51, DMC1, ZIP1 and RAD24, that represent different classes of recombination genes, in the suppression of ectopic meiotic recombination. Our results provide evidence for two types of ectopic recombination: one that occurs in wild-type cells and a second that occurs predominantly when the checkpoint pathway is inactivated. We found that RAD51 suppresses ectopic recombination, whereas RAD52 is critical for it to occur. Moreover, chromosome synapsis did not appear to be involved in the suppression of ectopic recombination.
Results
The checkpoint clamp loader RAD24 works in a common pathway with RAD51 to suppress ectopic recombination
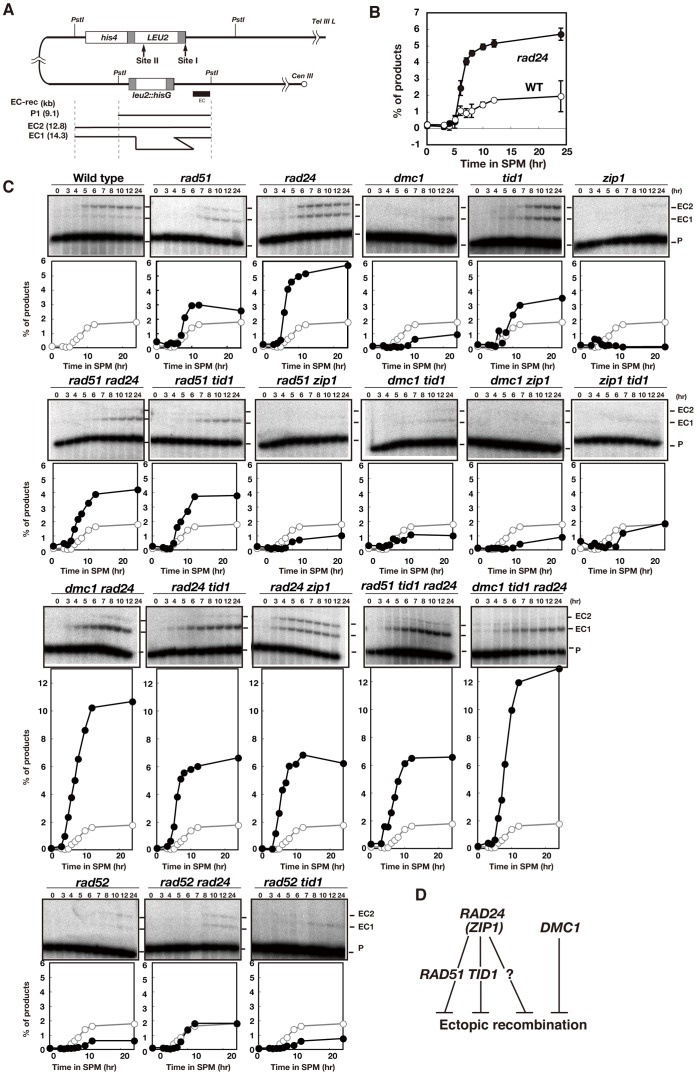
Previous studies by Bishop and colleagues showed that checkpoint mutants such as rad17, rad24, and mec1 increase ectopic recombination and that, when combined with dmc1 mutation, the mutants also show synergestic increase of ectopic recombination [45]. In current studies, we extended these observations by studying the effect of different combinations of mutations in genes involved in meiotic recombination, DNA damage response as well as chromosome synapsis. A genetic interaction occurs between DNA damage checkpoint genes and RAD51, both in mitosis and meiosis [46]. We followed recombination products/intermediates in a physical assay using the recombination hot-spot HIS4-LEU2 [53]. First, we measured products formed by ectopic recombination between the HIS4-LEU2 and leu2::hisG locus on chromosome III (Figure 1A).
Figure 1. Meiotic ectopic recombination in various mutants.
(A) Schematic representation of the HIS4-LEU2 and leu2::hisG loci. Homologous regions between the HIS4-LEU2 and leu2::hisG loci for ectopic recombination are shown in gray boxes. (B) Ectopic recombination in wild-type (WT) and rad24 mutant cells was analyzed by Southern blotting. Genomic DNA was digested with PstI. The percentage of ectopic recombination products (i.e., the relative amount of EC1 and EC2 bands) is shown. Error bars were obtained from three independent time courses. The error bars represent standard deviations (n = 3). Wild type, open circles; rad24 mutant, closed circles. (C) Ectopic recombination in various mutants was analyzed by Southern blotting. Quantification of products as in B is shown. For each strain, at least two independent time courses were performed. Representative data are presented for each mutant strain (closed circles); wild-type data (open circles) are described in each blot for comparison. (D) A model overview of pathways for the suppression of ectopic recombination. The “?” gene is a putative gene which may suppress ectopic recombination.
In the wild type, ectopic recombination products occurred at a frequency of 1.8% (Figure 1B and 1C). These recombination events were largely dependent on the meiosis-specific recA homolog DMC1 (Figure 1C). The frequency of ectopic products increased to 5.8% in the rad24 checkpoint mutant. These findings agree with a previous study [45]. Further, we found that a rad51 mutant also showed a modest increase in ectopic recombination to 3.0% (Figure 1C), suggesting a role for RAD51 in the suppression of ectopic recombination.
We also examined the relationship between checkpoint genes and the two RecA homologs RAD51 and DMC1 in ectopic recombination. The rad51 rad24 double mutant exhibited an intermediate level of ectopic recombination as compared with that of rad51 and rad24 single mutants (Figure 1C). Given that the level of ectopic products in the rad51 mutant was lower than that in the rad24 single mutant or the rad24 rad51 double mutant, the role of RAD24 in suppression of ectopic recombination has both RAD51-dependent and RAD51-independent components. In contrast, in the dmc1 rad24 double mutant there is a dramatic increase in ectopic recombination, up to 10.5% as shown previously [45], indicating that DMC1, unlike RAD51, can suppress ectopic events in the absence of RAD24.
TID1/RDH54 suppresses ectopic recombination
The DNA translocase Tid1/Rdh54 is required for normal meiotic recombination [54]. Tid1/Rdh54 promotes the recombination activity of both Rad51 and Dmc1 [29], [55]. When ectopic recombination was examined in the tid1/rdh54 null mutant, there was a modest increase in ectopic products (∼2-fold as compared with wild type), similar to that of rad51 (Figure 1C). Combining rad51 and tid1/rdh54 resulted in a higher level of ectopic products (1.3-fold) than that seen in the single mutants, suggesting differential contributions of RAD51 and TID1/RDH54 to suppression of ectopic recombination. Similar levels of ectopic recombination were observed in the rad24 single, tid1/rdh54 rad24 double and rad51 tid1/rdh54 rad24 triple mutants. Thus, it appears that rad24 suppression of ectopic recombination involves two additive components, a RAD51-dependent one and a TID1/RDH54-dependent one, which are partially overlapping functions. It should be noted that the level of ectopic recombination products in rad24 is somewhat higher than that in rad51 tid1/rdh54, suggesting that there may be a third pathway of suppression of ectopic recombination mediated by RAD24 (Figure 1D). These data suggest cooperation of RAD24, RAD51 and TID1/RDH54 in normal suppression of ectopic recombination. As in the case of the rad24 single mutant, introduction of dmc1 to the tid1/rdh54 rad24 double mutant increases the frequency of ectopic recombination from 6% to ∼12%. This reinforces the idea that DMC1, not RAD51 or its partner TID1/RDH54, plays a critical role in blocking ectopic events in the absence of RAD24.
RAD52 promotes ectopic recombination
Ectopic events occur even in the absence of the RecA homologs RAD51 or DMC1 (Figure 1C). Given that RAD52 can promote recombination in the absence of RAD51 in mitosis [56], [57], we also examined the role of RAD52 in ectopic recombination. The rad52 single mutant decreased ectopic recombination relative to that of wild type (2.7-fold; Figure 1C). Moreover, the combination of the rad24 or the tid1/rdh54 mutations with the rad52 mutation reduced ectopic recombinant formation as compared with that of the respective single mutants by 2.9- and 7.5-fold, respectively. Therefore, RAD52 plays a critical role in intra-chromosomal ectopic recombination in meiosis. However, the rad52 mutation did not completely abolish ectopic recombination in any of the three backgrounds, indicating that some ectopic events occur independently of RAD52 function. Therefore, most but not all ectopic events during meiosis depend on RAD52.
A ZMM gene, ZIP1, is required for ectopic recombination in wild-type, rad51 and tid1/rdh54 cells
ZMM genes, including ZIP1, promote CO formation as well as chromosome synapsis [13]. Like all members of the ZMM group, the zip1 mutant displays reduced CO levels, and increased NCO levels [13], [37], [38]. Without ZIP1, paired axial elements do not synapse [38]. We hypothesized that ectopic recombination levels might be restricted by assembly of the SC pairing interactions and therefore elevated in the zip1 mutant because of its synapsis defect. Contrary to this prediction, ectopic recombinant levels were markedly reduced to almost undetectable levels in the zip1 mutant (7.3-fold; Figure 1C), indicating that rather than repressing ectopic recombination, ZIP1 is required for it to occur. The negative effect of the zip1 mutation on ectopic recombination levels was largely epistatic to the positive effects of rad51 and tid1/rdh54 mutations; the rad51 zip1 and tid1/rdh54 zip1 double mutants showed 4.4- and 3.8-fold reductions, respectively, in ectopic products relative to levels in the respective single mutants (Figure 1C). Thus, ZIP1 appears to play a critical role in the formation of ectopic products in the wild-type contexts as well as in rad51 and tid1/rdh54 mutant contexts.
rad24 is epistatic to zip1 in meiotic recombination
In contrast to the situation in the wild type or in rad51 and tid1/rdh54 single mutants, addition of a zip1 mutation in the rad24 mutant did not alter the abnormally high levels of ectopic products (Figure 1C). This result indicates that there are two distinct pathways for ectopic recombination, one that predominates in wild-type cells and depends on ZIP1, and a second pathway seen in the rad24 mutant that is ZIP1-independent (Figure 1D). This is consistent with the recent finding that the Rad24 clamp loader is necessary for efficient loading of ZMM proteins onto meiotic chromosomes (MS & AS, unpublished results).
RAD51 and RAD24 suppress resection of DSB ends
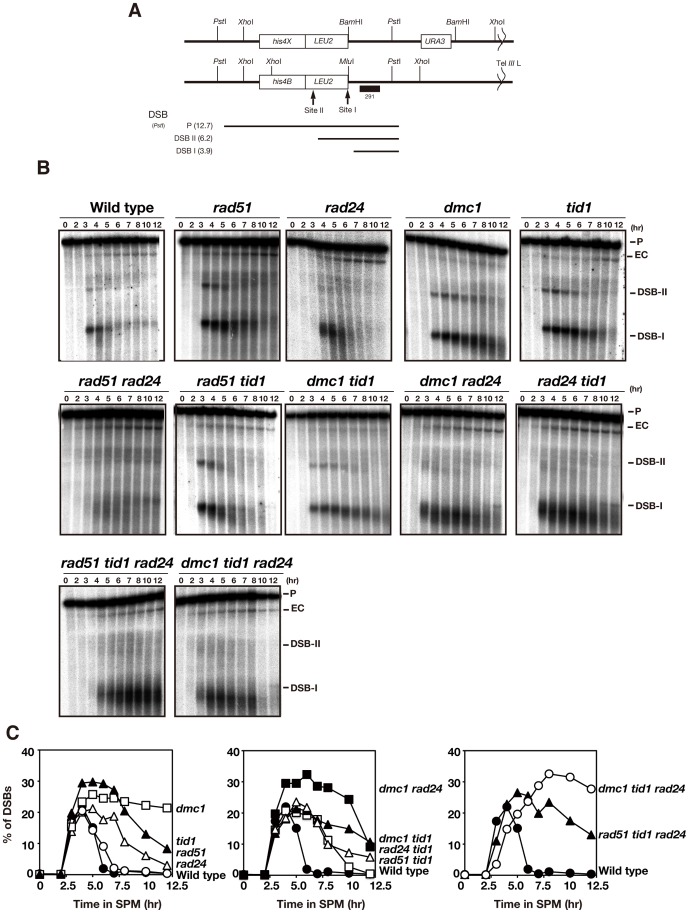
We also monitored DSB repair at the HIS4-LEU2 locus in above mutants (Figure 2A). All single, double and triple mutants showed delayed disappearance of DSBs (Figure 2B and 2C), indicating a defect in the repair of DSBs during meiosis. Most of the mutant combinations, particularly ones with the rad24 mutation, also showed elevated resection of the DSB ends relative to the wild type. The rad51 rad24 double mutant, in particular, exhibited particularly diffuse bands in the DSB regions, which are indicative of extensive nucleolytic degradation (Figure 2B). This suggests that RAD51 and RAD24 have redundant roles in the protection of DSB ends from nuclease attack during meiosis. A similar protective role for rad24 during mitosis has been reported [58].
Figure 2. DSB repair in various mutants.
(A) Schematic representation of HIS4-LEU2 locus for DSB detection. (B) DSB repair in wild-type and various mutant cells was analyzed by Southern blotting. Genomic DNAs were digested with PstI. For each strain, at least two independent time courses were performed. Representative data are shown for each strain. (C) Quantification of DSB at site I (B). The quantification of DSBs in the rad51 rad24 mutant is not shown due to smear nature of DSB bands. Left graph, wild type, closed circles; rad24, open circles; rad51, open triangles; tid1, closed triangles; dmc1, open rectangles. Middle graph, wild type, closed circles; rad24 tid1, open triangles; dmc1 tid1, closed triangles; rad51 tid1, open rectangles; rad24 dmc1, closed rectangles. Right graph, wild type, closed circles; dmc1 tid1 rad24, open circles; rad51 tid1 rad24, closed triangle.
Discussion
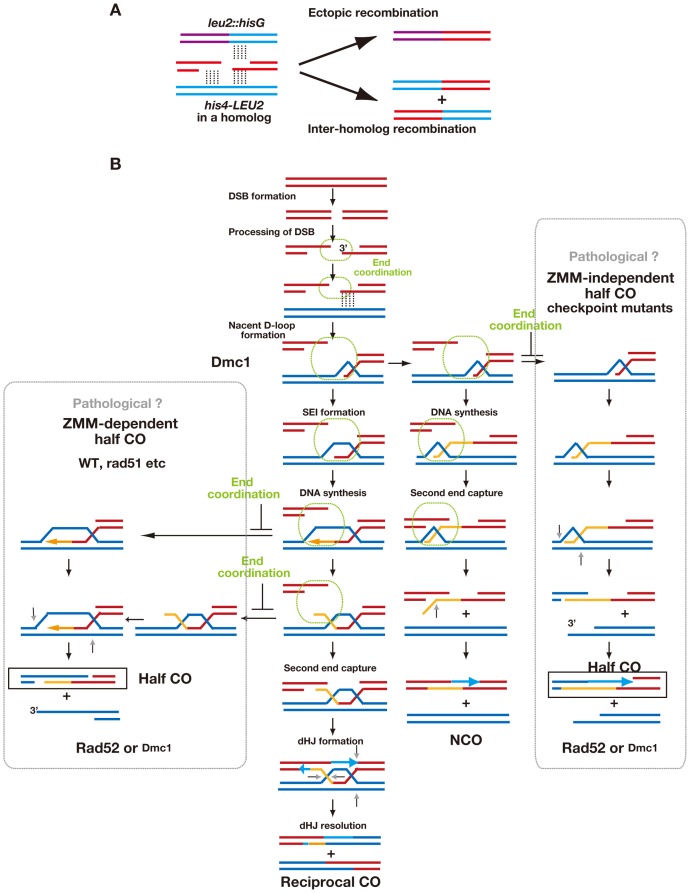
Meiotic recombination can produce two distinct products, COs and NCOs. These products are formed through different branches of the recombination pathway after the formation of DSBs (Figure 3). Meiotic COs are predominantly reciprocal between homologous chromosomes and are promoted by ZMM proteins. ZMM -dependent recombination intermediates appear to be critical for reciprocal CO formation, with coordinated direction of the two DSB ends to the same target for exchange [13]. By analyzing intra-chromosomal ectopic recombination, we were able to describe a novel pathway(s) for the formation of half-CO products. The exchange between LEU2 of HIS4-LEU2 and leu2::hisG, which was initiated at DSB site I [45], is forced to be non-reciprocal, as the homology is limited to one end of the DSB (LEU2 side; Figure 3A). Wild-type cells showed very low levels of ectopic recombination, whereas some mutants showed increased levels of these half-CO events. This suggests that targeting of the second DSB end to the same site as the first is regulated. A simple mechanism for end-coordination would be physical linkage of the two ends. Alternatively, interaction of the second end with the target could be indirectly regulated by interactions of the first end. For example, there might be a negative feedback mechanism to guarantee that both ends go to the same target DNA. This could occur by regulated asymmetric loading of recombination enzymes such that only one end of a DSB is endowed with strand invasion activity [9], [25], [28] or by specific disruption of interactions in which opposite DSB ends engage different targets [8], [59]. A non-exclusive alternative is that ectopic recombination between HIS4-LEU2 and leu2::hisG might be repressed because of structural constraints coupled with end coordination. For example, partner ends might be constrained to a local segment of the axial element [60], [61].
Figure 3. A model of meiotic recombination with pathological pathways.
(A) Only one side of DSB I in the HIS4-LEU2 locus has homology to leu2::hisG, which may lead to half-COs (top). A heterologous region of the leu2::hisG to LEU2 (left) is shown in purple. This is distinct from a canonical reciprocal CO pathway (bottom). (B) Two potential pathological pathways for ectopic recombination, which result in the formation of half-COs. Parental DNAs are shown in red or blue. Newly-synthesized strand is in orange. End-coordination is shown in green.
One interesting observation in this study is that ectopic recombination in some strains largely depended on the ZMM gene ZIP1. This ZIP1-dependent ectopic recombination seems to operate in wild-type, rad51, tid1/rdh54 and dmc1 cells. It is likely that ZIP1-dependent ectopic recombination is a pathological outcome that occurs when the normal end coordination mechanism fails (left pathway in Figure 3B). Because ZMM genes play a critical role in the formation of SEIs and dHJs [7], [13], it is likely that half-CO products in this pathway are formed via single-end interactions that yield SEIs or dHJs as intermediates.
In addition, there is a second pathway by which half-COs can occur during meiosis (pathway on the right in Figure 3B) that appears to be activated when checkpoint signaling is defective. Unlike the case in wild-type cells, ectopic recombinant formation in the checkpoint mutant, rad24, did not require ZMM function. Given that the checkpoint mutation was epistatic to the zip1 mutation with respect to meiotic recombination, this ZMM-independent ectopic recombination appears to reflect a pathway branch based on an earlier decision point than the normal ZMM-dependent recombination. In wild-type cells, the ZMM-dependent CO pathway follows the early CO/NCO decision with NCO pathway is branched as a default [7], [10]. Thus, it is likely that ZMM-independent ectopic products are a pathological derivative of the normal NCO pathway. Again we propose that this pathway is suppressed in the wild type by end-coordination functions. Our results strongly suggest that DSB end-coordination is key to the progression of the recombination complex to intermediates that are designated to become COs via the ZMM pathway.
Many of the requirements for ectopic recombination are different from those for allelic recombination. Two recA homologs, RAD51 and DMC1, are critical for inter-homolog recombination. The data in the paper show that RAD51 suppresses ectopic recombination in an otherwise wild-type strain whereas DMC1 only suppresses ectopic recombination in the rad24 background. Strand invasion in ectopic recombination seems to be carried out by Rad52 as well as Dmc1. Consistent with this, Rad52 catalyzes D-loop formation in vitro [62]. The role of Rad52 in half-CO formation might be similar to that in break-induced replication during mitosis [56]. Among proteins that suppress ectopic recombination, checkpoint proteins appear to play the most important role. The role of checkpoint proteins in the suppression does not appear to be related to their role in CO formation through the recruitment of ZMM proteins. Rather, the ability of checkpoint proteins to collaborate with Rad51/Rad54 for DNA repair processes [46] might be relevant.
It is widely believed that chromosome synapsis promotes recombination between homologous chromosomes. Earlier study on ectopic recombination suggests that interhomolog interactions restrict ectopic recombination [52]. We found that the zip1 mutant, which is defective in chromosome synapsis, reduces ectopic recombination. This suggests that chromosome synapsis may not be involved in the early steps of recombination, such as D-loop formation. Instead, Zip1 appears to be involved in stabilizing later recombination intermediates to promote CO.
Our physical study here as well as previous genetic studies on NAHR provided new insight on the ectopic recombination in meiosis [48]–[52]. However, it is important to point out that these studies used assays for ectopic recombination between sequences where the extent of homology is severely limited by nearby flanking heterology. We need to develop an assay to study ectopic recombination between sequences with long homology; e.g. recombination between sequences with more than 10kb homology on different chromosomal locations.
Our results showed that defects in the canonical meiotic recombination pathways lead to increased NAHR, which would presumably result in chromosome instability in gametes [4]. Therefore, genes involved in meiotic recombination may be a risk factor for genome instability.
Materials and Methods
Strains
All strains described here are derivatives of the S. cerevisiae SK1 diploid strain NKY1551 (MATα/MAT a , lys2/”, ura3/”, leu2::hisG/”, his4X-LEU2-URA3/his4B-LEU2, arg4-nsp/arg4-bgl). The genotypes of each strain used in this study are described in Table 1. The tid1/rdh54 null alleles were constructed by PCR-mediated gene disruption using KanMX6 genes as described [63].
Table 1. Strain list.
| Strain Number | Genotypes |
| NKY1551 | MATα/a , ho::LYS2/”, lys2/”, ura3/”, leu2::hisG/”, his4X-LEU2(BamHI)-URA3/his4B-LEU2(MluI), arg4-nsp/arg4-bgl |
| MSY2746 | NKY1551 with rad51::hisG-URA3-hisG |
| MSY717 | NKY1551 with rad24::LEU2 |
| MSY2237 | NKY1551 with dmc1::LEU2 |
| MSY1712 | NKY1551 with tid1/rdh54::KanMX6 |
| MSY2820 | NKY1551 with zip1::LEU2 |
| MSY2082 | NKY1551 with rad51::hisG-URA3-hisG, rad24::LEU2 |
| MSY2069 | NKY1551 with rad51::hisG-URA3-hisG, tid1/rdh54::KanMX6 |
| MSY2712 | NKY1551 with rad51::hisG-URA3-hisG, zip1::LEU2 |
| MSY219 | NKY1551 with dmc1::LEU2, tid1/rdh54::KanMX6 |
| MSY2719 | NKY1551 with dmc1::LEU2, zip1::LEU2 |
| MSY2135 | NKY1551 with tid1::KanMX6, zip1::LEU2 |
| MSY2096 | NKY1551 with dmc1::LEU2, rad24::LEU2 |
| MSY2103 | NKY1551 with rad24::LEU2, tid1/rdh54::KanMX6 |
| MSY2168 | NKY1551 with rad24::LEU2, zip1::LEU2 |
| MSY2122 | NKY1551 with rad51::hisG-URA3-hisG, tid1/rdh54::KanMX6, rad24::LEU2 |
| MSY2137 | NKY1551 with dmc1::LEU2, tid1/rdh54::KanMX6, rad24::LEU2 |
| MSY2777 | NKY1551 with rad52::hisG-URA3-hisG |
| MSY2822 | NKY1551 with rad52::hisG-URA3-hisG, rad24::LEU2 |
| MSY2825 | NKY1551 with rad52::hisG-URA3-hisG, tid1/rdh54::KanMX6 |
Analyses of meiotic recombination
Time course analyses of DNA events in meiosis and cell cycle progression were performed as described [29]. Ectopic recombination was analyzed as described [45]. Blots were scanned using a PhosphoImager, BAS3,000 (Fuji film) and quantified using ImageQuant (GE Healthcare) software. Ectopic recombination and DSB repair were analyzed more than twice and representative results are shown.
Acknowledgments
We thank Drs. Doug Bishop and Neil Hunter for discussion and Dr. Doug Bishop for careful editing. We are also indebted to members of the Shinohara lab for discussion.
Funding Statement
This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture to AS and MS, as well as grants from the Asahi-Glass Science Foundation, the Uehara Science Foundation, the Mochida Medical Science Foundation and the Takeda Science Foundation to AS. MS was supported by the Japanese Society for the Promotion of Science through the “Next Generation World-Leading Researchers” program, initiated by the Council for Science and Technology Policy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- 3. Goldfarb T, Lichten M (2010) Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sasaki M, Lange J, Keeney S (2010) Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kadyk LC, Hartwell LH (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae . Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keeney S (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53. [DOI] [PubMed] [Google Scholar]
- 7. Hunter N, Kleckner N (2001) The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- 8. Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, et al. (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N (2008) Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- 11. McMahill MS, Sham CW, Bishop DK (2007) Synthesis-dependent strand annealing in meiosis. PLoS Biol 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwacha A, Kleckner N (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- 13. Börner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- 14. Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- 15. Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470. [DOI] [PubMed] [Google Scholar]
- 16. Bishop DK (1994) RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092. [DOI] [PubMed] [Google Scholar]
- 17. Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK (1998) Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev 12: 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwacha A, Kleckner N (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- 19. Cloud V, Chan YL, Grubb J, Budke B, Bishop DK (2012) Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shinohara A, Ogawa T (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- 21. Sung P (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121. [DOI] [PubMed] [Google Scholar]
- 22.Sasanuma H, Tawaramoto SM, Lao JP, Hosaka H, Sanda E, et al.. (2013) A new protein complex promoting the assembly of Rad51 filaments. Nat Comms. in press. [DOI] [PMC free article] [PubMed]
- 23. Bishop DK, Zickler D (2004) Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- 24. Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, et al. (2004) A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119: 927–940. [DOI] [PubMed] [Google Scholar]
- 25. Kurzbauer MT, Uanschou C, Chen D, Schlogelhofer P (2012) The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24: 2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsubouchi H, Roeder GS (2004) The budding yeast Mei5 and Sae3 proteins act together with dmc1 during meiotic recombination. Genetics 168: 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petukhova G, Stratton S, Sung P (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393: 91–94. [DOI] [PubMed] [Google Scholar]
- 28. Shinohara M, Gasior SL, Bishop DK, Shinohara A (2000) Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc Natl Acad Sci U S A 97: 10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629. [DOI] [PubMed] [Google Scholar]
- 30. Agarwal S, Roeder GS (2000) Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- 31. Chua PR, Roeder GS (1998) Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- 32. Shinohara M, Oh SD, Hunter N, Shinohara A (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. [DOI] [PubMed] [Google Scholar]
- 33. Hollingsworth NM, Ponte L, Halsey C (1995) MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- 34. Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- 35. Ross-Macdonald P, Roeder GS (1994) Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- 36. Tsubouchi T, Zhao H, Roeder GS (2006) The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell 10: 809–819. [DOI] [PubMed] [Google Scholar]
- 37. Storlazzi A, Xu L, Schwacha A, Kleckner N (1996) Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci U S A 93: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sym M, Engebrecht JA, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- 39. Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- 40. Hochwagen A, Amon A (2006) Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol 16: R217–228. [DOI] [PubMed] [Google Scholar]
- 41. Majka J, Burgers PM (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A 100: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Majka J, Niedziela-Majka A, Burgers PM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
- 44. Lydall D, Nikolsky Y, Bishop DK, Weinert T (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843. [DOI] [PubMed] [Google Scholar]
- 45. Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, et al. (1999) Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shinohara M, Sakai K, Ogawa T, Shinohara A (2003) The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson DA, Stahl FW (1999) Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153: 621–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jinks-Robertson S, Petes TD (1985) High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc Natl Acad Sci U S A 82: 3350–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kupiec M, Petes TD (1988) Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae . Mol Cell Biol 8: 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lichten M, Borts RH, Haber JE (1987) Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae . Genetics 115: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goldman AS, Lichten M (1996) The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics 144: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goldman AS, Lichten M (2000) Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A 97: 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cao L, Alani E, Kleckner N (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae . Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- 54. Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, et al. (1997) Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klein HL (1997) RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, et al. (2001) RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev 15: 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Signon L, Malkova A, Naylor ML, Klein H, Haber JE (2001) Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 21: 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jessop L, Rockmill B, Roeder GS, Lichten M (2006) Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Blat Y, Protacio RU, Hunter N, Kleckner N (2002) Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- 61. Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, et al. (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T (2001) Homologous pairing promoted by the human Rad52 protein. J Biol Chem 276: 35201–35208. [DOI] [PubMed] [Google Scholar]
- 63. Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]