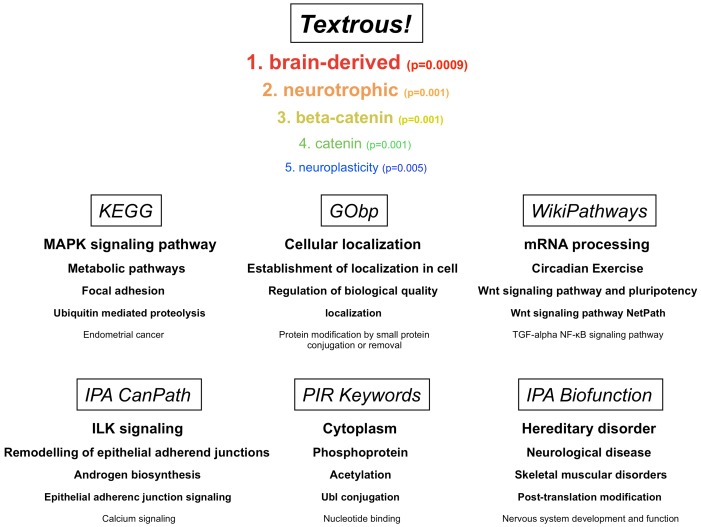
Figure 5. Multiple comparison of the functional accuracy and specificity of Textrous!-extracted data with other data analysis modules.
The top five most significantly associated words obtained from Textrous! collective analysis of the mouse learning dataset are compared to the top 5 most significantly enriched, KEGG pathways, GO-biological processes (GObp), WikiPathways, Ingenuity Pathway Analysis (IPA) Canonical Signaling Pathways (IPA CanPath), Protein Information Resource Keywords (PIR Keywords) and IPA BioFunctions generated using WebGestalt (KEGG, GObp, WikiPathways), IPA (CanPath, BioFunctions) and NIH-DAVID (PIR Keywords) respectively. The text size and descending sequential orientation indicate the first to the fifth most significantly enriched group for each analytical mode illustrated.