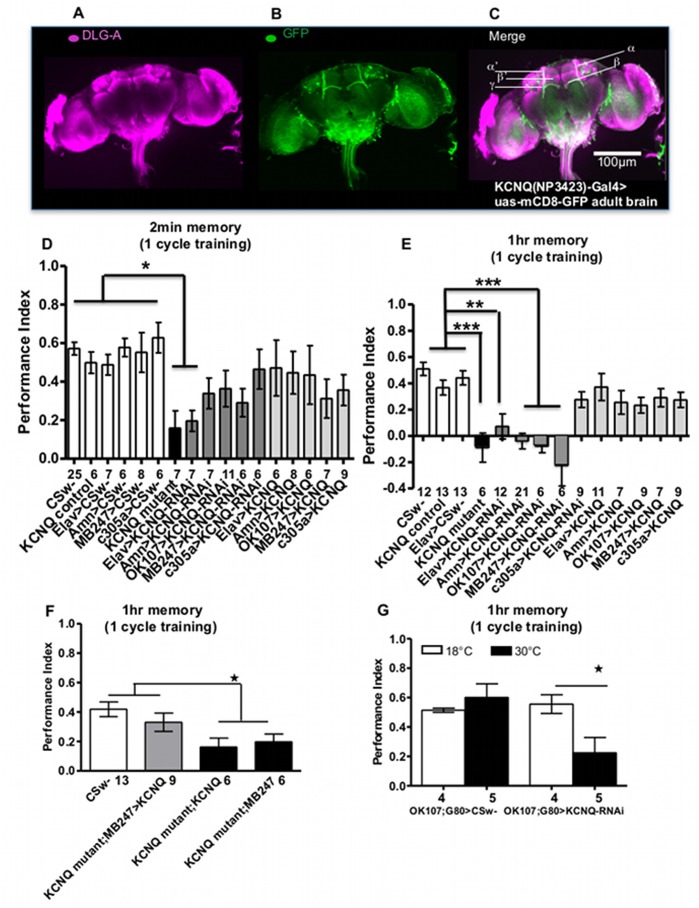
Figure 1. KCNQ signalling is required in the mushroom body α and β neurons for short-term memory.
A.–C. Adult brains containing a Gal4 enhancer trap (KCNQNP3423) in the KCNQ gene locus revealed broad neuronal expression of KCNQ (labelled by membrane bound GFP in green (B)) especially in the fly memory structures of the mushroom body α and β neurons and surrounding neurons known to be visualised by DLG-A (Ruiz-Cañada et al., 2002) staining (in magenta (A), co-localised structures in white (C)). D. Initial (2 min) memory was reduced in the KCNQ mutant (black bar) and flies with reduced KCNQ levels (dark grey bars) in all neurons (Elav-Gal4, uas-KCNQ-RNAi) (p<0.05) compared with controls (CSw-, KCNQ control, and Gal4, +, white bars) but did not lead to any change in memory (p>0.05) between the remaining genotypes. E. KCNQ mutants and flies with reduced KCNQ in the mushroom body (OK107-Gal4 or MB247-Gal4, uas-KCNQ-RNAi), DPM (amn-Gal4, uas-KCNQ-RNAi) (p<0.001) or all (Elav-Gal4, p<0.01) neurons have a significant reduction in 1 hr STM compared to controls (CSw-, KCNQ control and Gal4, +), while KCNQ overexpression (light grey bars) had no effect (p>0.05) with these promoters. F. Mushroom body α/β neuron expression of the KCNQ transgene in the KCNQ mutant background (KCNQ mutant; MB247-Gal4, uas-KCNQ) rescued the KCNQ mutant memory deficit with its memory being greater (p<0.05) than KCNQ mutant with Gal4 or uas alone (KCNQ mutant; MB247-Gal4 and KCNQ mutant; uas-KCNQ) but statistically indistinguishable (p>0.05) from control (CSw- wildtype) levels. Data in D-F were analysed by 1-way ANOVA with Bonferroni post-hoc test. G. 1 hr memory was measured in OK107-Gal4, Gal80ts, uas-KCNQ-RNAi and OK107-Gal4, Gal80ts, CSw- control flies raised at 18°C throughout development and then tested at 18°C conditions that prevent transgene expression (white bars). These scores were compared to the 1 hr memory of the same genotypes raised at 18°C throughout development and then shifted to 30°C allowing KCNQ transgene expression 2 days prior and during behavioural testing (black bars). 2-way ANOVA indicates significant differences due to interaction between temperature and genotype (p = 0.0195). Post-hoc analysis showed OK107-Gal4, Gal80ts, uas-KCNQ-RNAi had less (p<0.05) memory at 30°C compared to flies at 18°C flies (∼100 flies per n). In this and all subsequent figures, error bars represent SEM with no asterisk p>0.05, *p<0.05, **p<0.01 and ***p<0.001. n is denoted by the number between the x axis and genotype names with experiments performed on multiple different days (∼100 flies were used per n, unless otherwise stated).