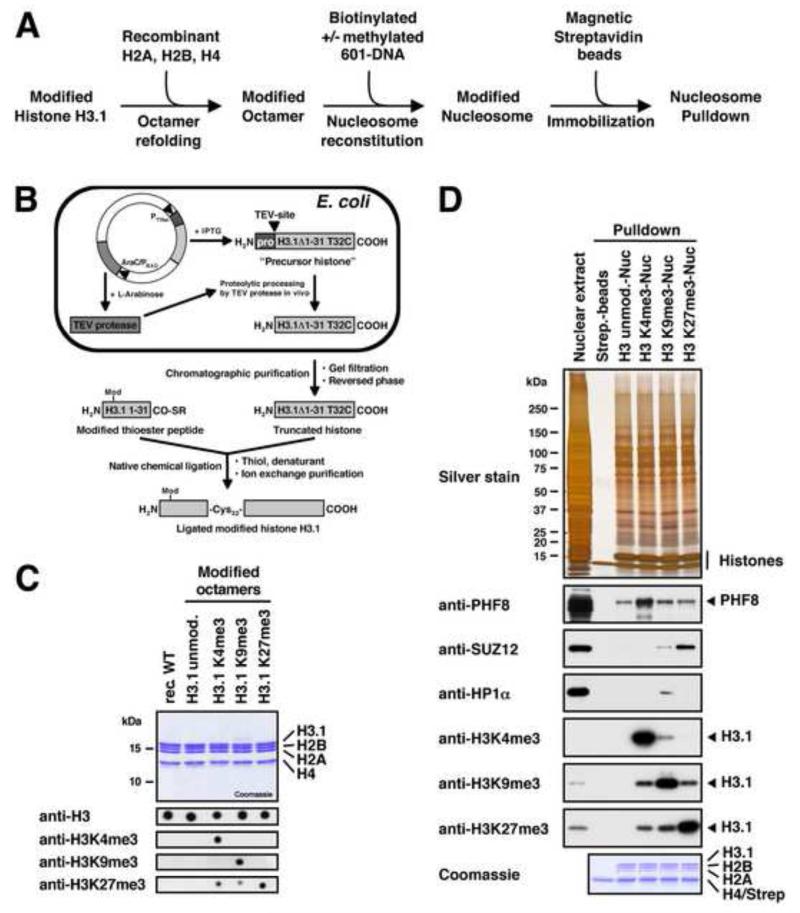
Figure 1. Preparation of Reconstituted Modified Nucleosomes.
(A) Experimental strategy for the preparation of immobilised and modified nucleosomes for pulldown studies. (B) The native chemical ligation strategy for generating post-translationally modified histone H3.1. We bacterially express an IPTG-inducible truncated histone precursor containing a modified TEV-cleavage site (ENLYFQ↓C) followed by the core sequence of histone H3.1 starting from glycine 33. The plasmid also contains TEV-protease under the control of the AraC/PBAD-promoter. TEV-protease accepts a cysteine instead of glycine or serine as the P1′-residue of its recognition site, and upon arabinose induction it processes the precursor histone into the truncated form (H3.1Δ1-31 T32C) which is purified and ligated to modified thioester peptides spanning the N-terminal residues 1 to 31 of histone H3.1. All ligated histones contain the desired modification and a T32C mutation. (C) Summary of the modified histone octamers. The upper panel shows 1 μg of each octamer separated by SDS-PAGE and stained with Coomassie. For the bottom panel octamers were dot blotted on PVDF-membranes and probed with modification-specific antibodies as indicated. The anti-H3K27me3 antibody shows slight cross-reactivity with H3K4me3 and H3K9me3. (D) Functional test of the nucleosome affinity matrix. R10K8-labelled nuclear extract was incubated with immobilised modified nucleosomes as indicated. Binding of PHF8, HP1α, and SUZ12 was detected by immunoblot. Equal loading was confirmed by silver and Coomassie staining. Modification of histone H3 was verified by immunoblot against H3 tri-methyl lysine marks. All three antibodies show slight cross-reactivity with the other histone marks. See also Figure S1.