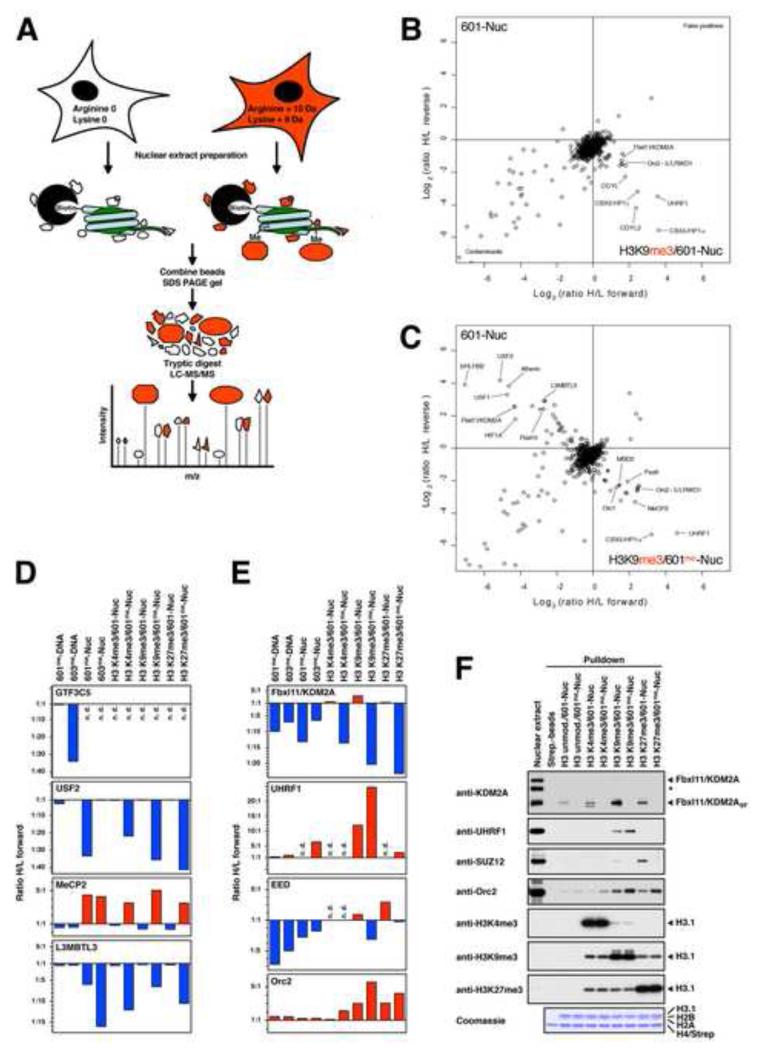
Figure 2. Identification of Nucleosome-interacting Proteins Regulated by DNA and Histone Methylation using SNAP.
(A) Experimental design of the SILAC nucleosome affinity purifications. Nuclear extracts are prepared from HeLaS3 cells grown in conventional “light” medium or medium containing stable isotope-labelled “heavy” amino acids. The resulting “light”- and “heavy”-labelled proteins can be distinguished and quantified by MS. Immobilised unmodified or modified nucleosomes are separately incubated with light or heavy extracts, respectively. Both pulldown reactions are pooled and eluted proteins are separated by SDS-PAGE. After in-gel trypsin digestion, peptides are analysed by high resolution MS. (B) Results of SNAP performed with H3K9me3-modified nucleosomes containing unmethylated 601-DNA. Shown are the Log2-values of the SILAC ratios (ratio H/L) of each identified protein for the forward (X-axis) and the reverse (Y-axis) experiments. The identities of several interacting proteins are indicated. Subunits of the MBD2/NuRD-complex are labelled in orange. (C) Results of SNAP performed with H3K9me3-modified nucleosomes containing CpG-methylated 601-DNA. For additional SNAP results see Figure S2 and Table S1. (D) Differential recognition of nucleosomes. The graphs show the forward SILAC enrichment values (Ratio H/L forward) of MeCP2, L3MBTL3, USF2, and the TFIIIC subunit GTF3C5 on CpG-methylated DNAs and modified nucleosomes. Binding to the modified nucleosomes or DNAs is indicated in red, exclusion is indicated in blue. If proteins were not detected (n.d.) no value is assigned. (E) Crosstalk between DNA and histone methylation. The graphs show the SILAC enrichment values of the proteins KDM2A, UHRF1, the PRC2 subunit EED, and the ORC subunit Orc2 as described in (D). (F) Immobilised modified nucleosomes were incubated with an independently prepared R0K0-nuclear extract as indicated. Binding of KDM2A, UHRF1, Orc2, and the PRC2 subunit SUZ12 was detected by immunoblot. Equal loading and modification of histone H3 were verified as in Figure 1D. The asterisk marks a cross-reactive band recognised by the KDM2A antibody.