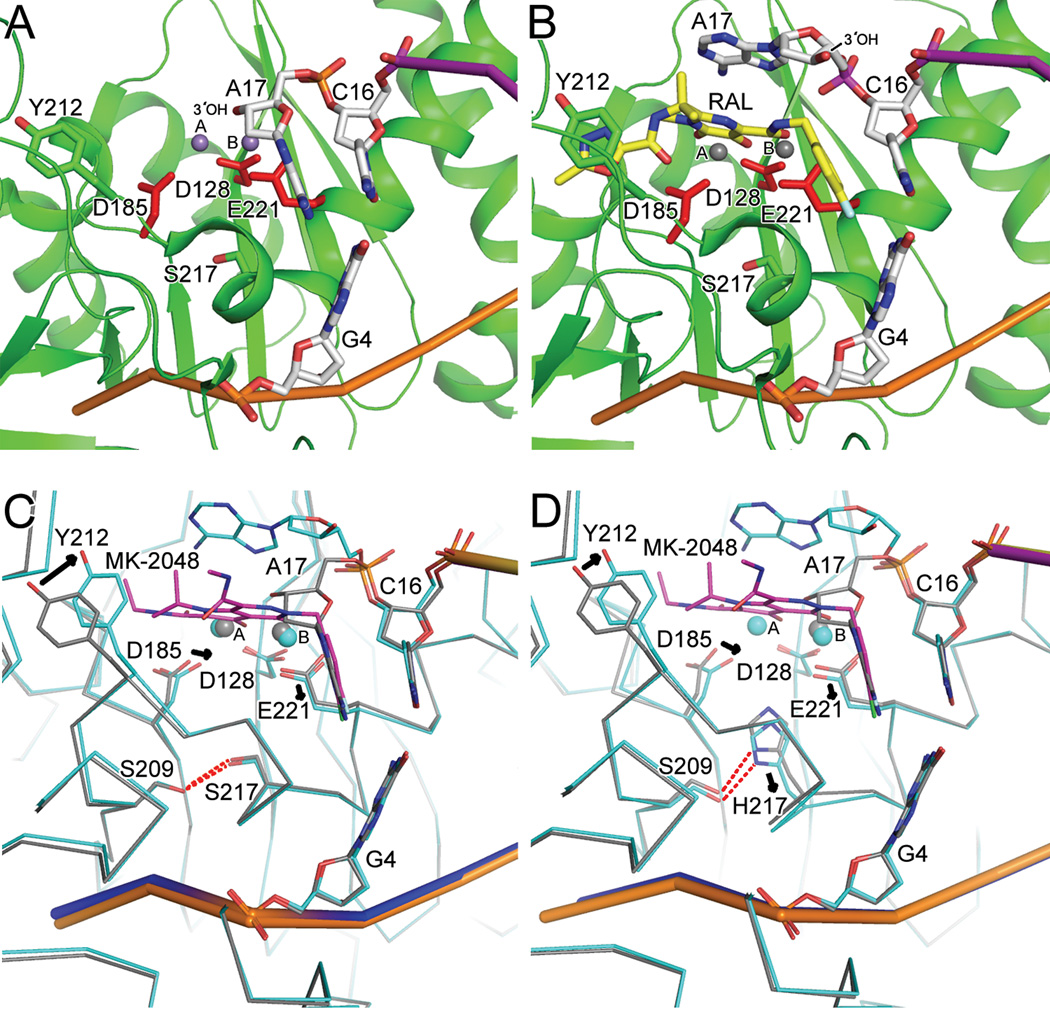
Fig. 6.
Wild type and S217H mutant intasome structures in committed and drug-bound forms elucidate the mechanism of INSTI action and basis for HIV-1 Q148H/R/K drug resistance. (A) Mn-bound active site (PDB code 3OY9) revealed the coordination of two metal ions by the DDE catalytic triad (see main text for additional details). (B) RAL (yellow scaffold) binding to an induced fit pocket formed through interactions with coordinated metal ion (Mg2+) and the penultimate C16/G4 bp of the vDNA end ejects the terminal adenine nucleotide (A17) and its affiliated 3′-OH nucleophile from the IN active site (PDB code 3OYA). (C) Superposition of wild-type intasome in committed (grey trace; same view as panel A) and MK-2048 (magenta backbone) bound (blue trace; PDB code 3OYB) conformations. (D) Comparison of MK-2048-bound (PDB code 3OYL; blue trace) and unbound S127H mutant (grey trace; PDB code 3OYK) intasome structures revealed significant active site conformational changes (alterations in side chain positions indicated by arrows) elicited by drug binding. Other labeling is same as in Figures 4 and 5.