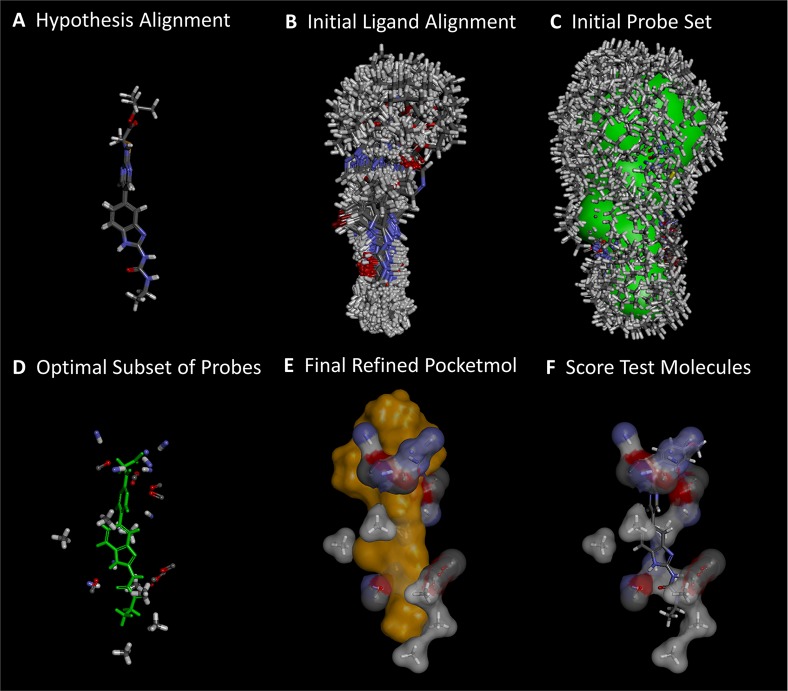
Figure 3.
Derivation and testing of a QMOD pocketmol proceeds in six automated steps: (A) an alignment seed hypothesis is constructed from 2 to 3 ligands; (B) 100–200 alignments for each training ligand are produced; (C) a large set of probes (many thousands) is created where interactions may exist; (D) a small near-optimal set is selected based on fit to experimental binding data and model parsimony; (E) probe positions and ligand poses are refined iteratively; (F) new molecules are tested by flexible alignment into the pocket to optimize score. The final pocketmol is used in a fixed configuration, but conformational flexibility within the corresponding protein pocket is represented by probes being places in multiple positions.