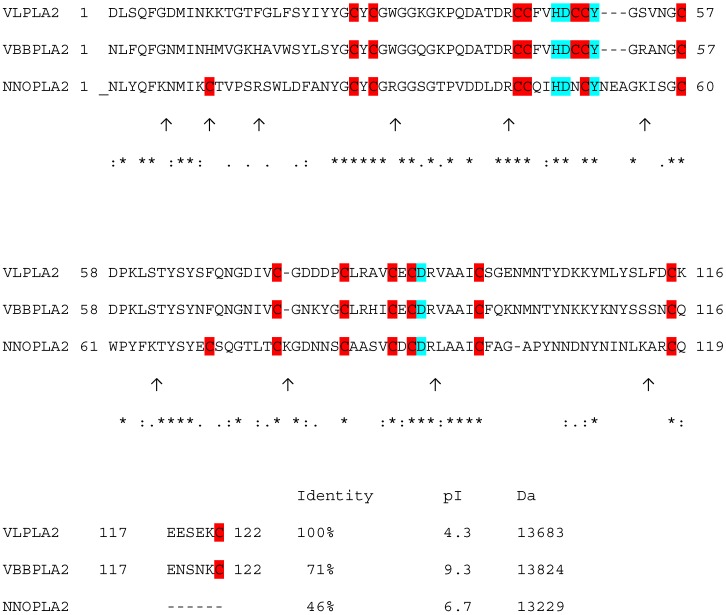
Figure 1.
Alignment of V. lebetina VLPLA2 (EU421953) [18], VBBPLA2 V. berus berus (P31854) [48] and NNOPLA2 isozyme E from N. naja oxiana (P25498) [49]. The alignment was performed using the program CLUSTAL W (1.83) multiple sequence alignment. “*” indicates positions which have a single, fully conserved residue; “:” indicates that one of the “strong” amino acid groups is fully conserved; “.” indicates that one of the “weaker” groups is fully conserved. Trypsin cleavage sites in NNOPLA2 are indicated as ↑. Cysteine residues are on red background, conserved catalytic network formed by four amino acid residues His48, Asp49, Tyr52 and Asp99 are on blue background.