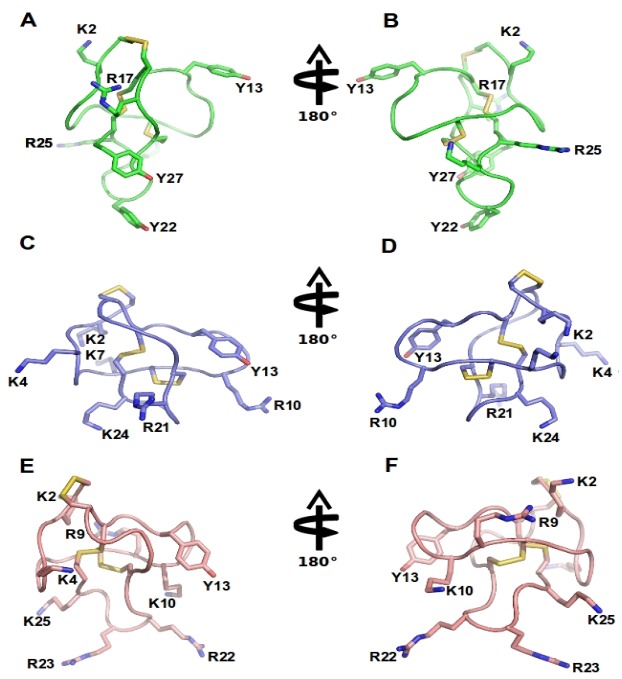
Figure 3.
ω-Conotoxins structure: NMR structure of GVIA (PDB 1TTL, green A-B), MVIIA (PDB 1 TTK, blue C-D) and MVIIC (PDB 1CNN, pink D-E). Represented are two different orientations. Disulfide bridges are shown in yellow and important amino acid residues, including Y13 (tyrosine13) and K2 (lysine2) and several positively charged residues exposed in the side chain are labeled.