Abstract
Many studies focused on the discovery of novel biomarkers for the diagnosis and treatment of disease states are facilitated by mass spectrometry based technology. HPLC coupled to mass spectrometry is widely used; miniaturization of this technique using nano-LC-MS usually results in better sensitivity, but is associated with limited repeatability. The recent introduction of chip-based technology has significantly improved the stability of nano-LC-MS, but no substantial studies to verify this have been performed.
To evaluate the temporal repeatability of chip-based nano-LC-MS analyses, N-glycans released from a serum sample were repeatedly analyzed using a nLC-PGC-chip-TOF-MS on three non-consecutive days. With an average inter-day CV of 4%, determined on log10 transformed integrals, the repeatability of the system is very high. Overall, chip-based nano-LC-MS appears a highly stable technology, which is suitable for profiling of large numbers of clinical samples for biomarker discovery.
Keywords: biomarker discovery, nano-LC-MS, Time-of-flight, N-glycans
INTRODUCTION
The development of biomarkers is important for the diagnosis and treatment of disease states and may serve several purposes, including the early detection of diseases (e.g. [1]), differentiation between disease categories [2] and to predict and monitor the response to treatment [3,4]. Therefore, numerous studies are being conducted aiming to identify such markers for various diseases at different levels in biology, including genomics [5], proteomics [6], metabolomics [7] and glycomics [8,9].
The technology used in proteomics, metabolomics and glycomics based studies often comprises liquid chromatography (LC) coupled to mass spectrometry (MS). This strategy is widely accepted, and has resulted in tremendous progress in our understanding of biochemical processes. Nano-scale LC-MS has several potential advantages such as the higher sensitivity, efficiency and resolution in separation, selectivity, and shorter analysis time. The use of nano-LC-MS is also associated with some limitations, including the relatively high LC back-pressure due to smaller particles for separation, lower stability of the LC-gradient and, most importantly, decreased stability of the electrospray ionization process resulting in less consistent data. This has resulted in the somewhat slower adoption of nano-LC-MS in the field of biomarker discovery.
Recent technological developments, including but not limited to the introduction of microfluidic chip-based LC-MS, have significantly improved repeatability of nano-LC-MS analyses, both with respect to retention time and spray conditions. These developments allow the use of nano-LC-MS for the determination of compounds associated with a disease as suitable biomarkers for the clinical setting. Our group was the first to introduce chip-based nano-LC coupled to nano-ESI-TOF-MS for the analysis of N-glycans derived from human serum [10–12] and more recently blood [13].
Good stability and repeatability of the analytical system is essential for the development of clinical biomarkers [14]. However, the suitability of the microfluidic chip based nano-LC-MS technology has not been rigorously evaluated for use in large biomarker studies that span several days needed to analyze large number of samples. In this study we evaluated the temporal stability of the nLC-chip-TOF instrument as an analytical tool for the identification and validation of glycan biomarker candidates. N-glycans were released from one standard serum sample and purified using porous graphitized carbon (PGC) SPE. Stability of the nLC-chip-TOF-MS was assessed by the repeated analysis of a single sample over a period of three non-consecutive days.
MATERIALS AND METHODS
The methods used are described briefly here; more details of materials and methods can be found in the Electronic Supplementary Material.
N-Glycan sample preparation
N-Glycans were released from the proteins in 50 μl of serum standard (Sigma-aldrich, St.Louis, MO) as previously described [15]. Briefly, proteins were denatured and N-glycans were enzymatically released in a microwave. Graphitized carbon SPE was used to purify the N-glycans as previously described [15]. The sample was dried in vacuo prior to analysis.
Study design
The dry N-glycan sample was reconstituted in 225 μL of water and transferred to an injection vial for analysis; 1 μL of sample was used for injection. The same N-glycan sample was used for all three days, and stored at −80°C between the days. The N-glycan sample was analyzed continuously throughout a day with intermittent blanks on three non-consecutive days. On each day, we continuously injected 1 μL of sample as long as the instrument allowed during daytime in a real life setting, thus the number of samples analyzed varied by day: Twelve analyses were performed on Day 1, twenty-two analyses were performed on Day 2 and seventeen analyses were performed on Day 3; the analyses on days 1 and 2 were performed on a previously used chip with approximately 300 hours of operation, while for the analyses on Day 3 a new analytical chip was used.
nHPLC-chip-TOF-MS analysis
N-Glycans were analyzed using an Agilent nanoHPLC-chip-TOF-MS, in which a PGC chip II (Agilent technologies, Santa Clara, CA) packed with porous graphitized carbon was used. Upon injection, the sample was loaded onto the enrichment column and subsequently separated on the analytical column using a gradient of 3% ACN with 0.5% FA (solvent A) to 90% ACN with 0.5% FA (solvent B). The mass spectrometer was operated in positive mode.
Data analysis and statistics
Data analysis was performed using Masshunter® qualitative analysis according to Hua et al. [16] with modifications. Data was loaded into Masshunter® qualitative analysis, and glycan features were identified and integrated using the Molecular Feature Extractor algorithm. A retrosynthetic theoretical glycan library [17] containing 331 possible N-glycan compositions was used for glycan identification. For statistical analysis, the integral values were log10 transformed to reduce the influence of extreme values to meet the normality assumption. Patterns in total glycan area and the proportion of non-detectables over time were evaluated graphically as was the relationship with the proportion of non-detectables and area values. Coefficients of variance (CVs) were calculated to assess inter- and intra-day variability. All data are reported as mean ± SD unless specified otherwise.
RESULTS
N-Glycans from human serum, which consist of hexoses (H) N-acetylhexosamines (N), fucoses (F) and N-acetylneuraminic acid (S), were separated, identified and quantified using nLC-PGC-chip-TOF-MS. To separate the glycans, a PGC (5μ) stationary phase was used, which was previously shown to result in very good glycan separation [18]. It was chosen to leave the reducing end of the glycans intact, which results in the chromatographic separation of the anomers on the PGC stationary phase.
The experiments were performed on three non-consecutive days over a 10-day period. The first two days were performed using one chip, while the third day was performed using a different LC chip to determine the variations between microchips. The stability of the system as well as the separation of isomers is demonstrated in Figure 1, where the extracted ion chromatograms are overlayed for all the days' runs for four different glycan compositions: H7N2 at m/z 780.28, H3N4F1 at m/z 732.28, H4N4F1 at m/z 813.31 and H5N4S1 at m/z 966.85. For composition H3N4F1, which is expected to have only one isomer in serum, the anomer separation is clearly visible. All other compositions show more than two peaks and thus comprise anomers as well as other isomers. To further illustrate the stability of the system, mass spectra were extracted for two N-glycan structures (H4N4F1 at m/z 813.31 and 8.2 min. and H5N4S1 at m/z 966.85 and 9.5 min.) for six runs on the three different days; the spectra are shown in the Electronic Supplementary Material, Figure S1. The resulting mass spectra are very similar, not only in terms of ions detected, but also in terms of ion counts. Overall, this result shows that the elution patterns of individual glycans are highly reproducible with respect to retention times and peak areas within each single day of analysis. Between days, slight differences in the relative areas are observed. The largest was for H5N4S1 at m/z 966.85, however the deviation was within the acceptable range of the coefficient of variation (CV) of <10%.
Figure 1. Overlays of extracted ion chromatograms for four different glycan compositions.
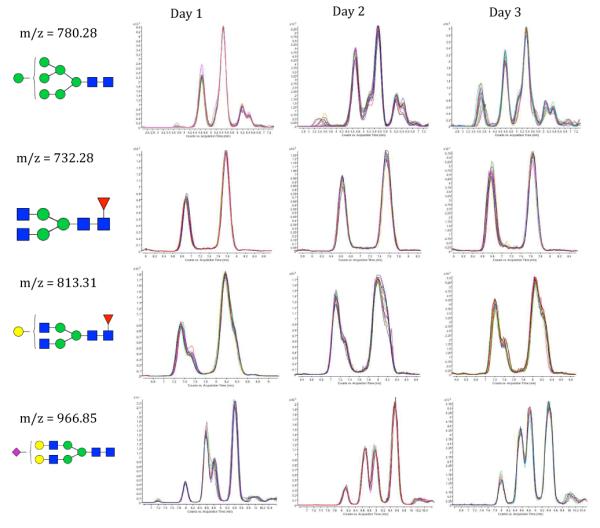
The extracted ion chromatograms for four different glycan compositions (H7N2 at m/z 780.28, H3N4F1 at m/z 732.28, H4N4F1 at m/z 813.31 and H5N4S1 at m/z 966.85, from top to bottom), stratified per day. For each day of analysis, an overlay of 10 chromatograms is depicted. Glycan compositions are given in terms of Hexose (H), N-acetylhexosamine (N), Fucose (F) and N-acetylneuraminic acid (S).
The experiments provide an opportunity to determine the consistency of the glycan signals in the nano-LC-MS method. Figure 2 illustrates the relationship between the proportion of the glycans not detected and the log of the mean peak area. Glycan compositions with absolute intensities above 105 (Log 5 with Log 8 maximum) are observed in every spectra. Mean log peak areas below 4.5 are observed in about 50% of the chromatograms, while those below 4.0 are observed less than 20% of the chromatogram. The results suggest a dynamic range of nearly four orders of magnitude (log 4 to log 8) for this method for the abundant peaks for consistent detection, while those near or less than log 4 are not consistently observed.
Figure 2. Relationship between the proportion of samples in which a glycan was not detected and the mean area of the glycan when detected.
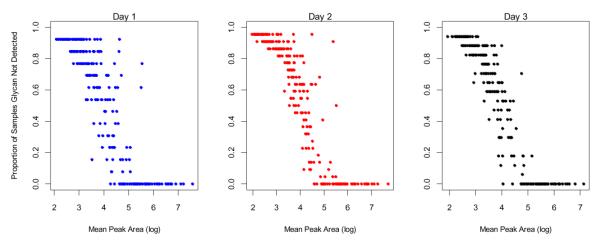
The results are separated for each of the three different days. The mean area values are log10 transformed. Each dot represents a glycan compound.
To evaluate the temporal stability of the method, the summed area of all glycan signals in each run was determined in sequence over the three day period (Figure 3). The overall glycan areas varied little within a day, however slightly lower glycan abundances were observed on Day 3 relative to Day 1 and 2 (5.76 × 107 compared to 1.40 × 108 and 1.62 × 108 for days 3, 1 and 2 respectively). The drop in intensities is likely due to the performance of the new chip. Day 1 and Day 2 were performed with the same chip, while a different chip was used on day 3. Another potential reasons for the decrease in the intensities for day 3 may be degradation of the oligosaccharides or absorption onto the vial surface. However, both are gradual processes that are expected occur across the three days. The first two days are relatively constant, while the drop is largest in Day 3 suggesting that the decrease is intrinsic to the chip and not due to sample loss. These experiments suggest that for consistency, particularly in biomarker studies, all samples should be performed on the same chip.
Figure 3. Repeatability of the analysis of serum-derived N-glycans using nLC-PGC-chip-TOF-MS.
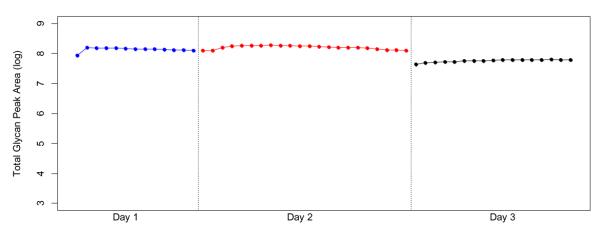
Each dot represents a total glycan area of all glycans in each run of the three days. Glycan areas are on log10 scale; Day 1 and day 2 were run on a chip with over 300 hours of operation, while day 3 was run on a new chip. Day 1 is represented by blue dots, day 2 by red dots and day 3 by black dots.
We further evaluated the stability of the nLC-chip-TOF instrument at the level of individual glycan compositions. Because this study is focused on repeatability, estimating or `imputing' missing values could affect the results. We therefore limited our analyses to the glycan compositions that were identified in all runs for each day. All glycan compositions that were not included for further data analysis were of low abundance, with integrals at an average of 3.5 × 10−4 relative to the highest glycan, as discussed previously. The intra-day variability was then assessed using 63, 58 and 57 glycan compositions that were consistently detected on day 1, 2 and 3, respectively. Using these glycans, the correlations in peak areas were calculated across runs within a day. For the correlation coefficients ranged (0.90 to 0.99) for Day 1 (0.70–0.99) Day 2, and (0.96–0.99) Day 3, exhibiting excellent within-day variability is generally very good. The lowest variation was obtained on day 3, when the new chip was used.
To evaluate the inter-day and intra-day repeatability of the instrument with respect to quantitation, we examined the areas of the 50 glycan compositions (listed in the Electronic Supplementary Material, Table S1) that were consistently detected in all 51 analyses performed throughout all three days. The coefficients of variation of the log10 transformed areas were calculated within a day and across days. The CVs were generally very small, with an average intra-day CV of 2.1% ± 1.7% and average inter-day CV of 3.9% ± 1.4%. Additionally, the inter- and intra-day CVs declined as the mean intensity level increased (i.e., lower abundance compounds were more variable across runs), however all CVs were less than 4%. The results indicate excellent repeatability of the glycan quantitation using nLC-chip-TOF instrumentation even for low abundance glycan compositions. We calculated the correlations for glycan compositions between Day 1 and 2, Day 1 and 3 and Day 2 and 3 (Electronic Supplementary Material, Table S2). Interestingly, the glycan correlation between Days 1 and 3 and between Days 2 and 3 was much larger than that between Days 1 and 2, indicating that the lower intensities observed on Day 3 did not alter the glycosylation patterns.
DISCUSSION
For biomarker discovery and development it is essential to have a stable analytical platform. To our best knowledge, this study represents the first comprehensive analysis of the temporal stability of nano-LC-chip-TOF-MS for the profiling of N-glycans.
The repeatability of the method was excellent, with the average intra-day CV of 2.1% ± 1.7% and average inter-day CV of 3.9% ± 1.4%. While it was observed that the incidence of detection of a certain glycan depends on its abundance, the overall glycan integration did not depend on the number of glycans detected in a given sample. These results indicate that shifts in total abundances of a run do not necessarily affect the general N-glycan profile. Variation appears to occur primarily between days, when the chip is removed and reinserted, and when a new chip is used. When a chip is removed and reinserted, the ionization environment within the source may alter slightly but sufficiently to affect the total ion abundances.
Although the repeatability was generally excellent, slightly better performance was obtained on day 3, where a new chip was used. The new column resulted in slightly more consistent retention times compared to the other two days where the chip had already logged hundreds of hours. In sample sets where hundreds of samples are analyzed, then a single chip may be sufficient to perform the analysis, while samples that number in the thousand may require some variation in the strategy so that variability between the beginning and the end can be minimized
While we showed high analytical repeatability of the nLC-chip-TOF-MS, there are other technical aspects that could influence the success of biomarker discovery studies. These include the effects of sample preparation itself, as well as storage of the samples prior to, during and after sample preparation [19]. These aspects, which are applicable not only to glycomics but also other `omics' such as proteomics or metabolomics in which mass spectrometry is applied, should be taken into account in design and interpretation of MS-based glycomics studies for biomarker discovery. We have performed systematic studies to determine the effects of sample preparation on the analytical repeatability using MALDI-FTICR-MS [15], and optimized the procedure for N-glycan release and purification from serum accordingly.
In conclusion, the repeatability of N-glycan profiles as analyzed by our platform was shown to be very high even when a combination of “old” and “new” chips are used. This study indicates that chip-based nano LC-MS technology is a very stable technology, which is suitable for biomarker discovery studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful for the funding provided by the National Institutes of Health (R21 CA135240, HD061923, HD059127, R01 GM049077, and UL1 TR 000002), the Department of Defense (CDMRP LCRP W81XWH1010635), the Tobacco Related Disease Research Program and the LUNGevity Foundation.
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
REFERENCES
- 1.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101(6):374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13(5):477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 3.Fine BM, Amler L. Predictive biomarkers in the development of oncology drugs: a therapeutic industry perspective. Clin Pharmacol Ther. 2009;85(5):535–538. doi: 10.1038/clpt.2009.9. [DOI] [PubMed] [Google Scholar]
- 4.Furihata T, Sawada T, Kita J, Iso Y, Kato M, Rokkaku K, Shimoda M, Kubota K. Serum alpha-fetoprotein level per tumor volume reflects prognosis in patients with hepatocellular carcinoma after curative hepatectomy. Hepatogastroenterology. 2008;55(86–87):1705–1709. [PubMed] [Google Scholar]
- 5.Ju W, Smith S, Kretzler M. Genomic biomarkers for chronic kidney disease. Transl Res. 2012;159(4):290–302. doi: 10.1016/j.trsl.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, Wong CH, Zhang Q, Chin A, Park KS, Goodman G, Gazdar AF, Sage J, Dinulescu DM, Kucherlapati R, Depinho RA, Kemp CJ, Varmus HE, Hanash SM. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;20(3):289–299. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishur RJ, Rea SL. Applications of mass spectrometry to metabolomics and metabonomics: detection of biomarkers of aging and of age-related diseases. Mass Spectrom Rev. 2012;31(1):70–95. doi: 10.1002/mas.20338. [DOI] [PubMed] [Google Scholar]
- 8.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820(9):1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Lebrilla CB, An HJ. The prospects of glycan biomarkers for the diagnosis of diseases. Molecular Biosystems. 2009;5(1):17–20. doi: 10.1039/b811781k. [DOI] [PubMed] [Google Scholar]
- 10.Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11(3):1958–1968. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alley WR, Jr., Madera M, Mechref Y, Novotny MV. Chip-based reversed-phase liquid chromatography-mass spectrometry of permethylated N-linked glycans: a potential methodology for cancer-biomarker discovery. Anal Chem. 2010;82(12):5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruhaak LR, Miyamoto S, Kelly K, Lebrilla CB. N-glycan profiling of dried blood spots. Anal Chem. 2012;84(1):396–402. doi: 10.1021/ac202775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kocher T, Pichler P, Swart R, Mechtler K. Quality control in LC-MS/MS. Proteomics. 2011;11(6):1026–1030. doi: 10.1002/pmic.201000578. [DOI] [PubMed] [Google Scholar]
- 15.Kronewitter SR, de Leoz ML, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. Human serum processing and analysis methods for rapid and reproducible N-glycan mass profiling. J Proteome Res. 2010;9(10):4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 136. 2011;18:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9(11):2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruhaak LR, Deelder AM, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2009;394(1):163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 19.Gast MC, van Gils CH, Wessels LF, Harris N, Bonfrer JM, Rutgers EJ, Schellens JH, Beijnen JH. Influence of sample storage duration on serum protein profiles assessed by surface-enhanced laser desorption/ionisation time-of-flight mass spectrometry (SELDI-TOF MS) Clin Chem Lab Med. 2009;47(6):694–705. doi: 10.1515/CCLM.2009.151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


