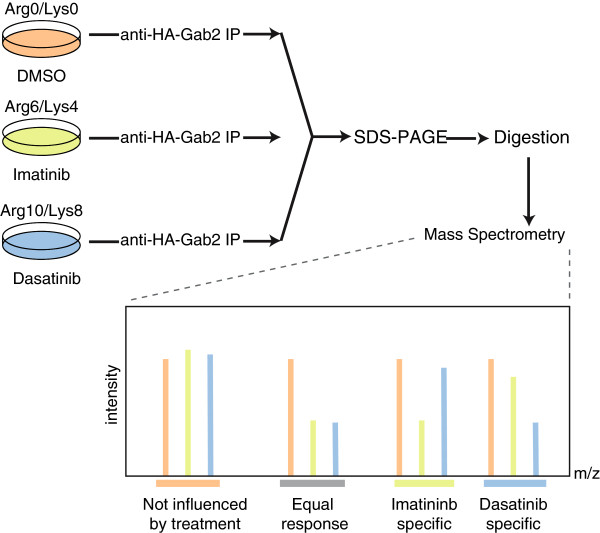
Figure 2.
Quantitative proteomics workflow. Differentially SILAC labeled K562 cells were treated for 4 h with either 1 μM imatinib or 0.01 μM and 1 μM dasatinib, and DMSO as control, respectively. Cells were lysed, Gab2 protein complexes purified by anti-HA sepharose, complexes eluted and combined 1:1:1 for further analysis. Proteins were separated by SDS-PAGE, in-gel digested using trypsin, and resulting peptide mixtures analyzed by LC-MS/MS. A biological replicate with reversed labels was performed. Specific Gab2 interactors and proteins binding unspecific to beads and antibody will be present in a 1:1:1 ratio. Protein interactions dependent on inhibitor sensitive phosphorylation sites will be reduced.