Abstract
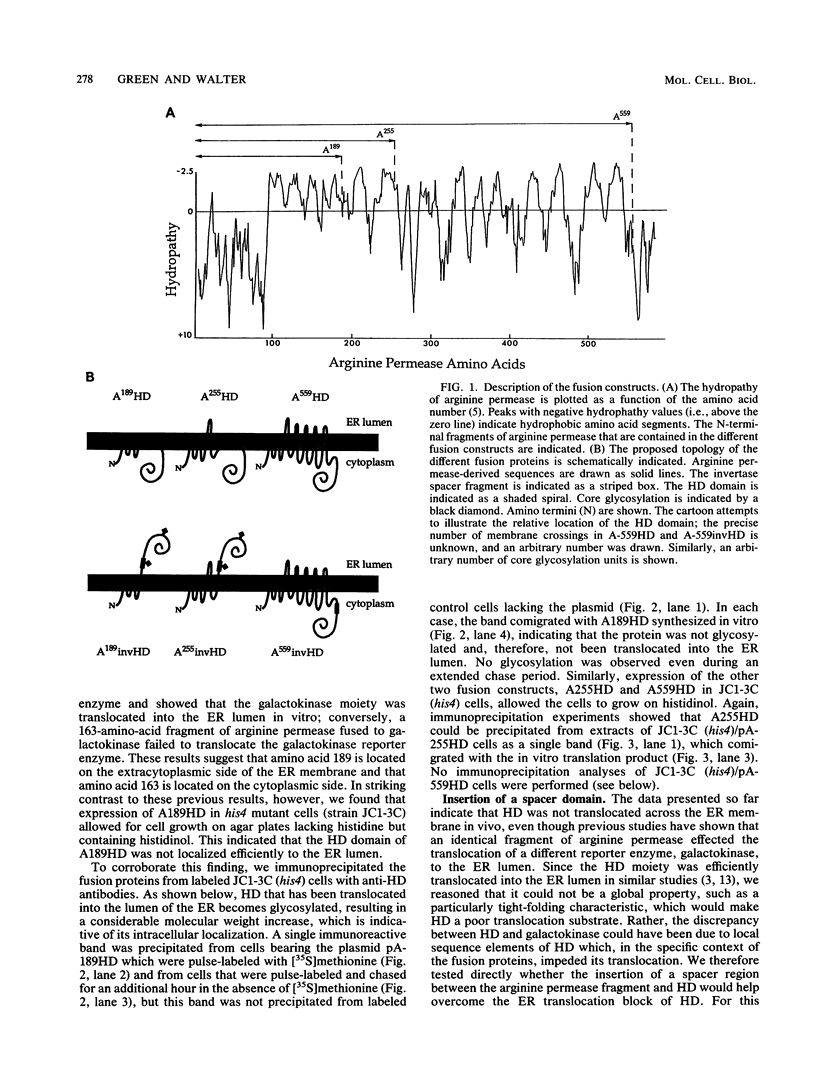
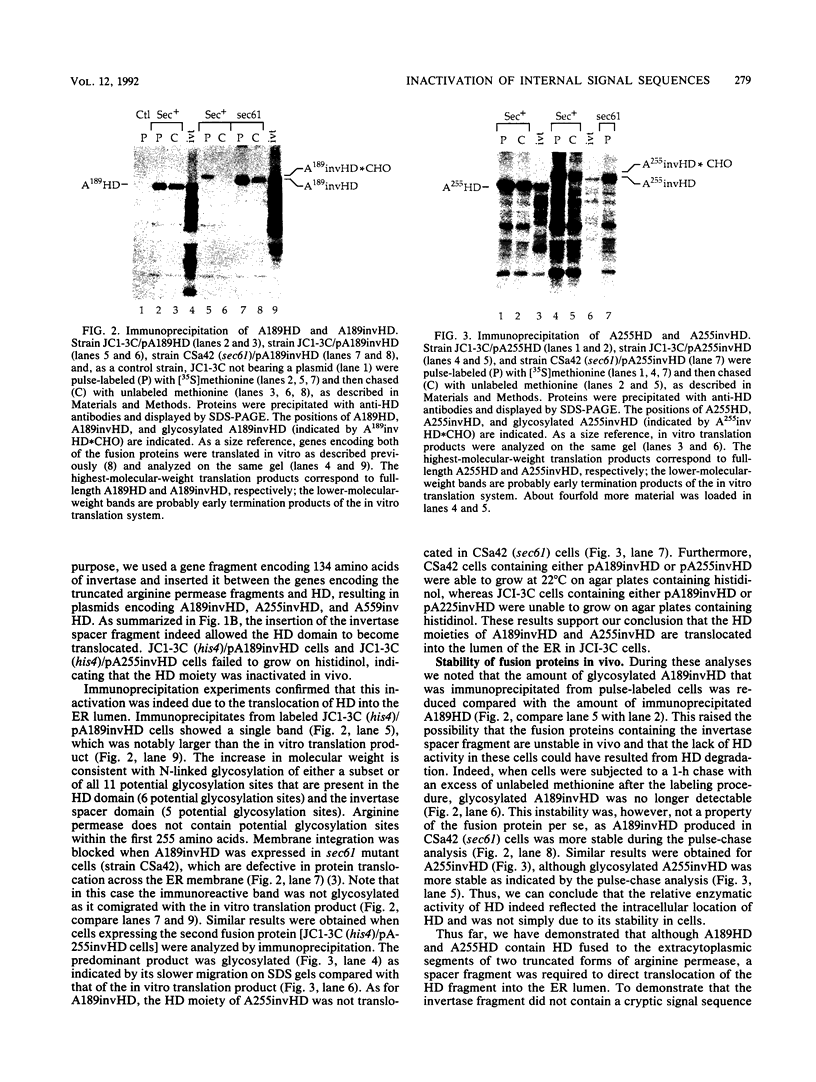
We have constructed three gene fusions that encode portions of a membrane protein, arginine permease, fused to a reporter domain, the cytoplasmic enzyme histidinol dehydrogenase (HD), located at the C-terminal end. These fusion proteins contain at least one of the internal signal sequences of arginine permease. When the fusion proteins were expressed in Saccharomyces cerevisiae and inserted into the endoplasmic reticulum (ER), two of the fusion proteins placed HD on the luminal side of the ER membrane, but only when a piece of DNA encoding a spacer protein segment was inserted into the fusion joint. The third fusion protein, with or without the spacer included, placed HD on the cytoplasmic side of the membrane. These results suggest that (i) sequences C-terminal to the internal signal sequence can inhibit membrane insertion and (ii) HD requires a preceding spacer segment to be translocated across the ER membrane.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Boyd D., Beckwith J. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9446–9450. doi: 10.1073/pnas.86.23.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987 Aug;105(2):633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Froshauer S., Green G. N., Boyd D., McGovern K., Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol. 1988 Apr 5;200(3):501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- Green G. N., Hansen W., Walter P. The use of gene-fusions to determine membrane protein topology in Saccharomyces cerevisiae. J Cell Sci Suppl. 1989;11:109–113. doi: 10.1242/jcs.1989.supplement_11.9. [DOI] [PubMed] [Google Scholar]
- Hansen W., Garcia P. D., Walter P. In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent posttranslational translocation of the prepro-alpha-factor. Cell. 1986 May 9;45(3):397–406. doi: 10.1016/0092-8674(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T. A., Lodish H. F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem. 1985 Sep 25;260(21):11831–11837. [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Sengstag C., Stirling C., Schekman R., Rine J. Genetic and biochemical evaluation of eucaryotic membrane protein topology: multiple transmembrane domains of Saccharomyces cerevisiae 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Cell Biol. 1990 Feb;10(2):672–680. doi: 10.1128/mcb.10.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988 Jul 1;174(4):671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G., Wickner W., Dalbey R. E. The cytoplasmic domain of Escherichia coli leader peptidase is a "translocation poison" sequence. Proc Natl Acad Sci U S A. 1988 May;85(10):3363–3366. doi: 10.1073/pnas.85.10.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]