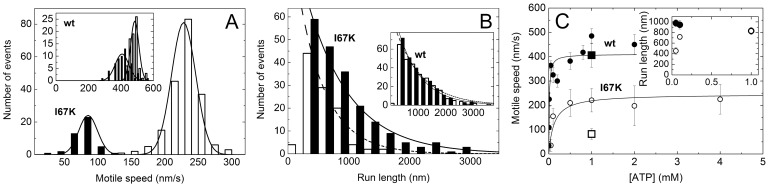
Figure 5. I67K mutation slows down and sensitizes processive motility.
A, In vitro motile speeds of wt-m5aHMM (inset) and I67K-m5aHMM (main panel) at 1 mM ATP, measured in actin gliding (solid columns) and single-molecule TIRF (open columns) assays. Gaussian fits yielded average speeds shown in C. B, Run lengths of wt-m5aHMM (inset) and I67K-m5aHMM (main panel) at 50 µM (open columns) and 1 mM (solid columns) ATP, determined in single-molecule TIRF experiments. Single exponential fits at 50 µM and 1 mM ATP (dashed and solid lines, respectively) yielded mean run lengths of 980±60 nm (wt-m5aS1, 50 µM ATP), 830±50 nm (wt-m5aS1, 1 mM ATP), 450±60 nm (I67K-m5aHMM, 50 µM ATP) and 830±70 nm (I67K-m5aHMM, 1 mM ATP) (see also panel C). Data points below 250 nm were omitted from fits. C, ATP concentration dependence of average speeds (main panel) and mean run lengths (inset) of wt-m5aHMM (solid symbols) and I67K-m5aHMM (open symbols), measured in single-molecule TIRF (circles) and actin gliding (squares, main panel, 1 mM ATP) assays. Hyperbolic fits to the TIRF speed datasets (main panel) yielded maximal speeds (v max) and half-saturating ATP concentrations (K ATP) listed in Table 1 . At 1 mM ATP, the run lengths (inset) of wt-m5aHMM and I67K-HMM were identical and the data points are indistinguishable. Error bars represent Gaussian half-widths (main panel) or standard errors of the fits (inset).