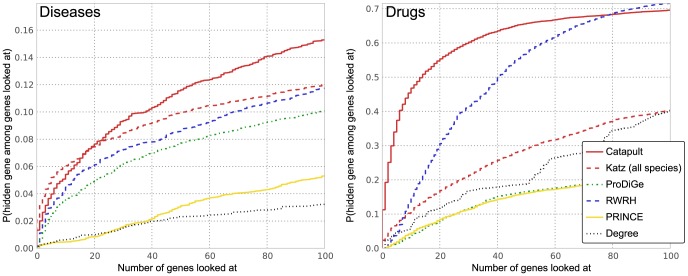
Figure 4. Comparison only using HumanNet.
Empirical cumulative distribution function for the rank of the withheld gene under cross-validation. Left panel corresponds to evaluation of OMIM phenotypes, and the right corresponds to drug data. The vertical axis shows the probability that a true gene association is retrieved in the top-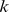 predictions for a disease. Katz and Catapult methods use all species information, and all the methods use the HumanNet gene network. PRINCE and RWRH methods are implemented as proposed in [7] and [8] respectively, but using HumanNet. ProDiGe method is implemented as discussed in Methods section. Again, as in Figure 3, Catapult (solid red) does the best. An important observation to be made from the plots is that PRINCE and RWRH methods perform relatively much better than in Figure 3, where HPRD network was used. (Note that there is no change to the ProDiGe, Katz and Catapult methods; they have identical settings as in Figure 3).
predictions for a disease. Katz and Catapult methods use all species information, and all the methods use the HumanNet gene network. PRINCE and RWRH methods are implemented as proposed in [7] and [8] respectively, but using HumanNet. ProDiGe method is implemented as discussed in Methods section. Again, as in Figure 3, Catapult (solid red) does the best. An important observation to be made from the plots is that PRINCE and RWRH methods perform relatively much better than in Figure 3, where HPRD network was used. (Note that there is no change to the ProDiGe, Katz and Catapult methods; they have identical settings as in Figure 3).